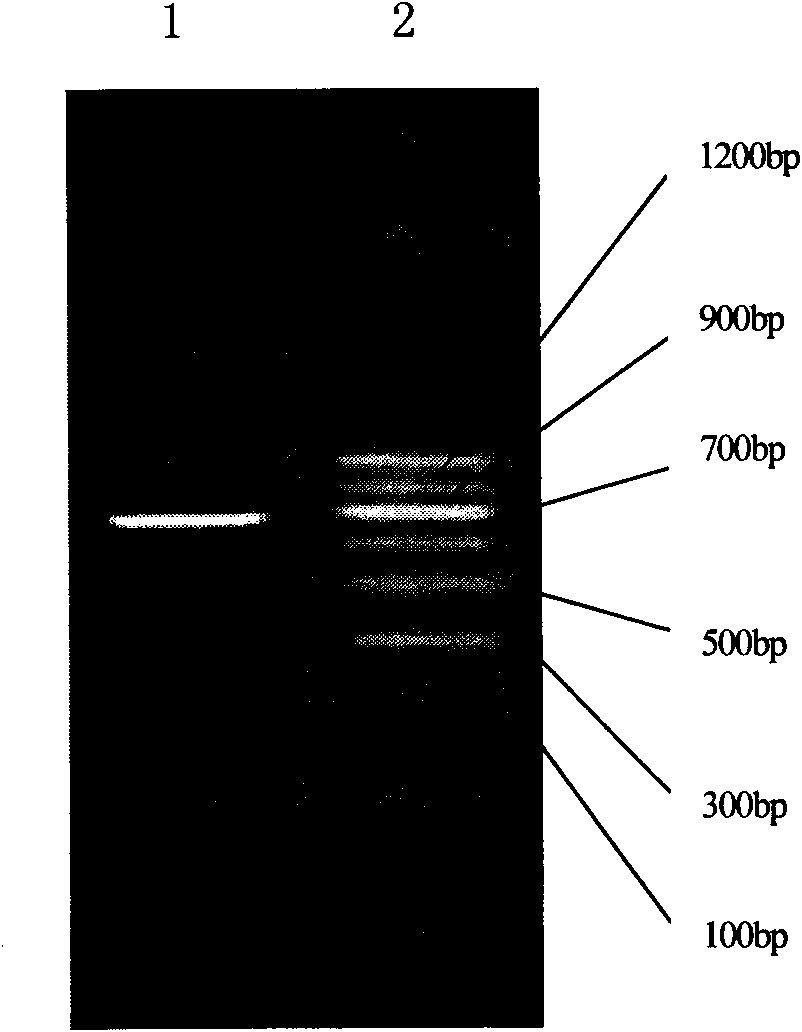
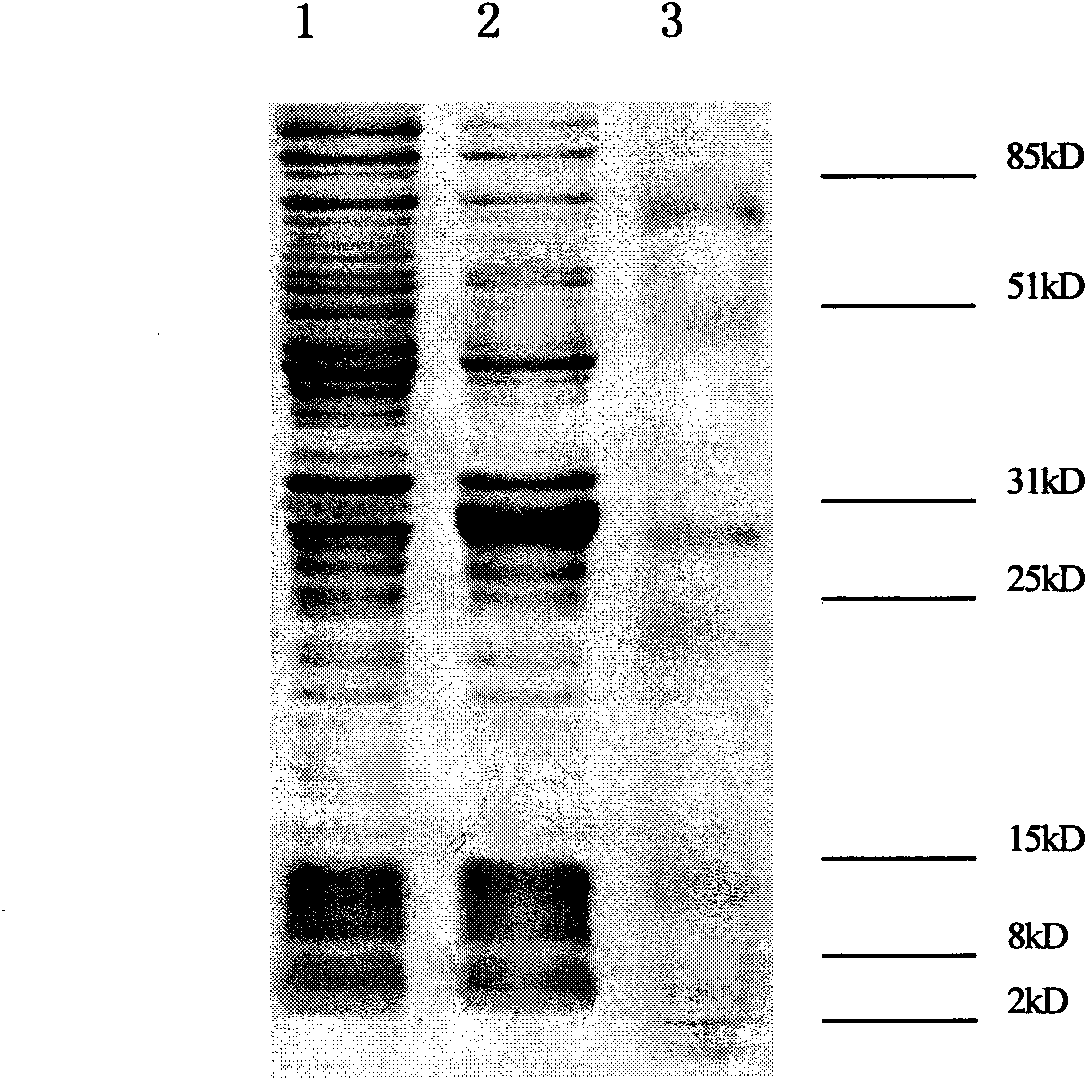
HIV-1 integrase expression bacterial strain and integrase inhibitor in-vitro screening model
An HIV-1 and integrase technology, applied in the field of genetic engineering, can solve the problems of unstable experimental results, environmental hazards, and high costs, and achieve the effects of simple and easy operation, low cost, and few interference factors for screening models.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Embodiment 1: The specific construction steps of the screening model provided by the present invention are as follows:
[0073] 1. Primer synthesis
[0074] Primers were designed according to the integrase gene sequence (from plasmid pNL4-3, available in NCBI):
[0075] His-Primer1: 5'-CAGCATATGTTTTTAGATGGAATAGATAAGGCCCAAGATGAAC-3'
[0076] NdeI restriction site: CATATG; start codon: ATG
[0077] Primer2: 5'-ATAGGATCCTTACTAATCCTCATCCTGTCTACTTGCCACAGAATCATCACCTGCC-3'
[0078] BamHI restriction site: GGATCC, stop codon: TTA, CTA; mutation site: AGA (to obtain C280S);
[0079] Primer3: 5'-CCACAATAAAAAAAGAAAAGGGGGGATTGGG-3'
[0080] Mutation site: AAA (get F185K);
[0081] Primer4: 5'-CCCAATCCCCCCTTTTTCTTTTTTTATTGTGG-3'
[0082] Mutation site: TTT;
[0083] LTR-Primer1: 5'-CAGTGGATCCGACTGTGGAAGGGCTAATTTGGTC-3'
[0084] BamHI restriction site: GGATCC; integrase action site: ACTG;
[0085] LTR-Primer2: 5'-CGACGAATTCGACTGCTAGAGATTTCCAC-3'
[0086] EcoRI restriction s...
Embodiment 2
[0111] Example 2: Construction of an in vitro screening model for HIV-1 integrase inhibitors
[0112] 1. Take out the constructed expression strain BL21-ET28a-IN stored in a glycerol tube from -80°C, inoculate it in 20ml of LB medium containing kanamycin sulfate 60μg / ml at a ratio of 1:1000, at 37°C, 180 rpm, overnight culture;
[0113] 2. The seed bacteria cultivated overnight in the previous step were inoculated into 10 bottles of 200ml LB medium according to 1:100, added kanamycin sulfate to a final concentration of 60μg / ml, 37°C, 180 rpm, and cultivated for 3 hours;
[0114] 3. Add the inducer IPTG to each bottle of culture in step 2 to a final concentration of 0.4mM, 37°C, 150 rpm, and induce expression for 3 hours;
[0115]4. Centrifuge each bottle of culture in step 3 at 5000rpm for 30 minutes to collect the bacteria, dissolve the bacteria in 100ml of 0.01M PBS buffer, freeze at -20°C, and then thaw;
[0116] 5. Ultrasonicate each bottle of culture in step 4 at 300W f...
Embodiment 3
[0120] Example 3: Application of HIV-1 integrase inhibitor in vitro screening model
[0121] 1. Add 20 pmol of the integrase obtained in Example 1, 200 ng pUC19-HIVLTR, 2 mM MnCl, 5 mM β-mercaptoethanol and different concentrations of samples to be tested in 10 μl of 20 mM Tris-HCl reaction system;
[0122] 2. After incubating the reaction system in step 1 at 37°C for 30 minutes, add 1 μl of 10×loading buffer to terminate the reaction, and run 1% agarose gel electrophoresis at 60V for 20 minutes;
[0123] 3. Use the gel imaging system UVP Biolmaging System to observe the plasmid bands in the 2-step electrophoresis, and use LabWorks45Image Acquisition and Analysis Software 4.5.00.0 to analyze the IOD values of supercoiled, linear and open-circular DNA. The results are shown in Table 2. Using this model to detect, the integrase inhibitor standard can show the activity of integrase inhibition, and the result shows that the construction of the model is successful. Using this m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com