Screening system of HIV infected cell and applications thereof
A cell line and cell technology, which is applied to a screening system for HIV-infected cells and its application field, can solve the problems of time-consuming, lengthy operation steps, and the screening time needs to be improved, and achieves the effects of short detection time and easy operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
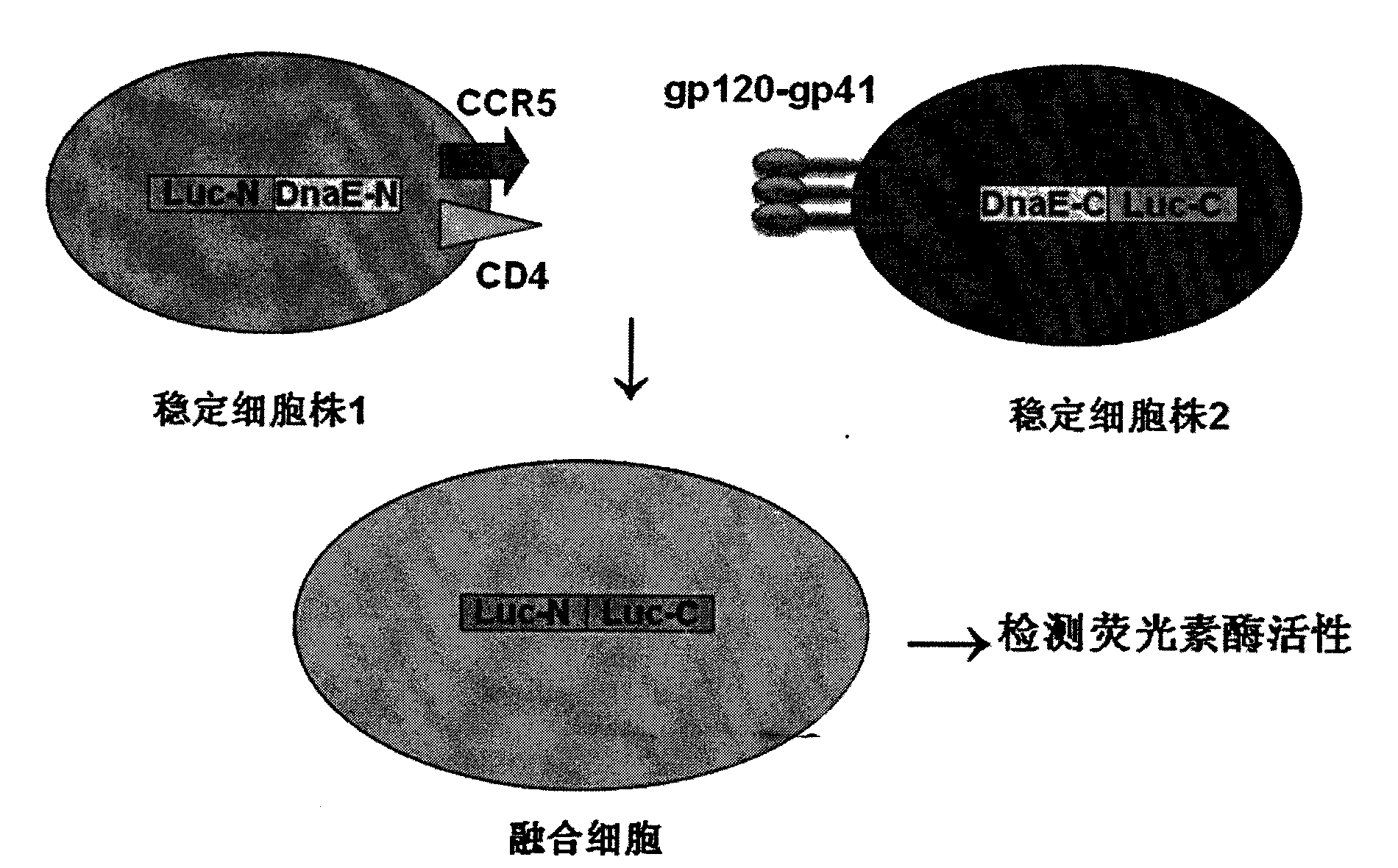
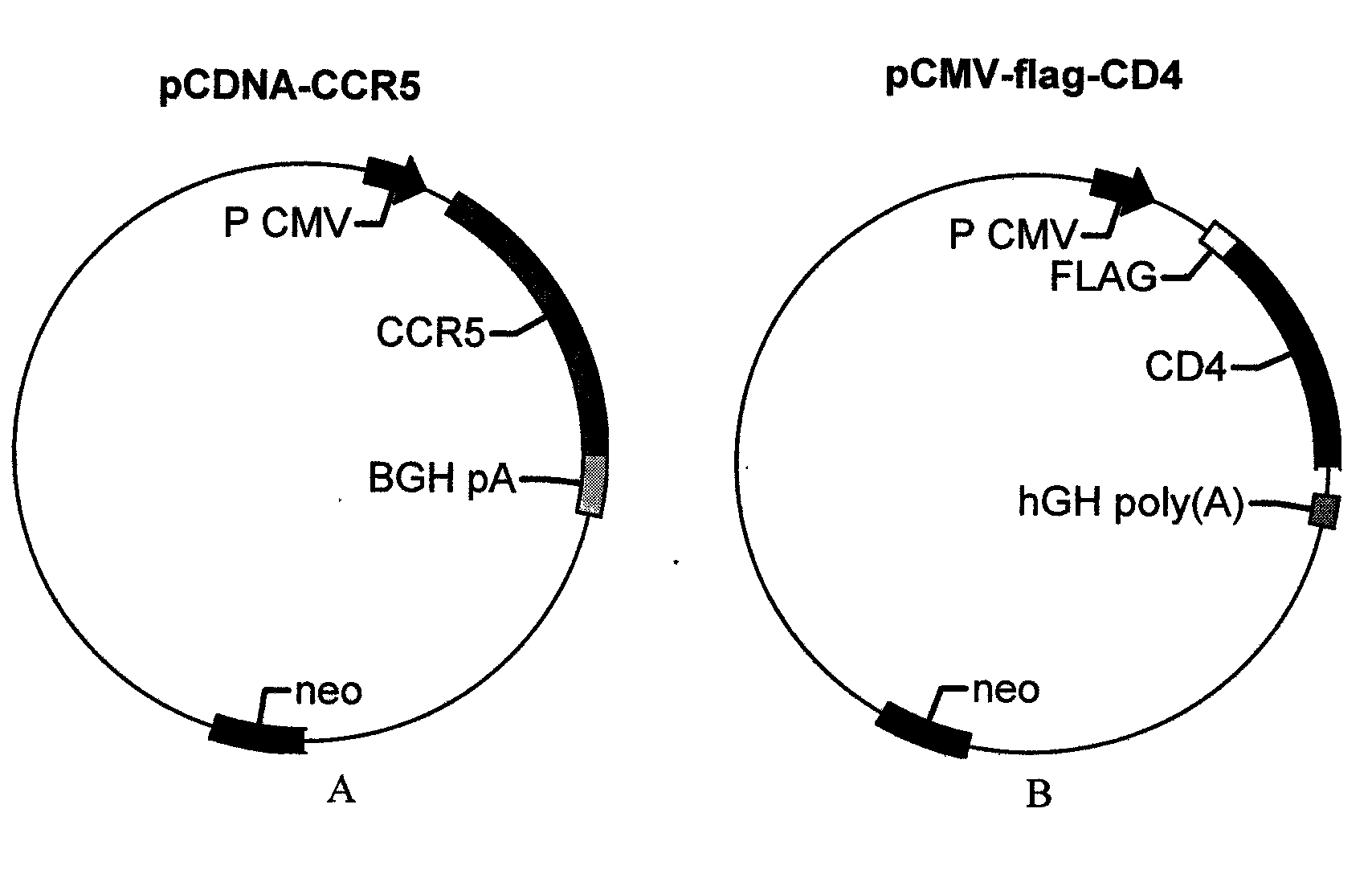
[0029] (1) Cloning the N-terminal (dnae-N) and C-terminal (dnae-C) of the DnaE intein gene from the genomic DNA of the cyanobacterium strain Anacystis nidulans R2 (PCC7942); cloning the CCR5 gene from the human macrophage genome (see SEQ ID NO.11 for the nucleotide sequence, and SEQ ID NO.12 for the amino acid sequence), CD4 gene (see SEQ ID NO.15 for the nucleotide sequence, and SEQ ID NO.16 for the amino acid sequence);
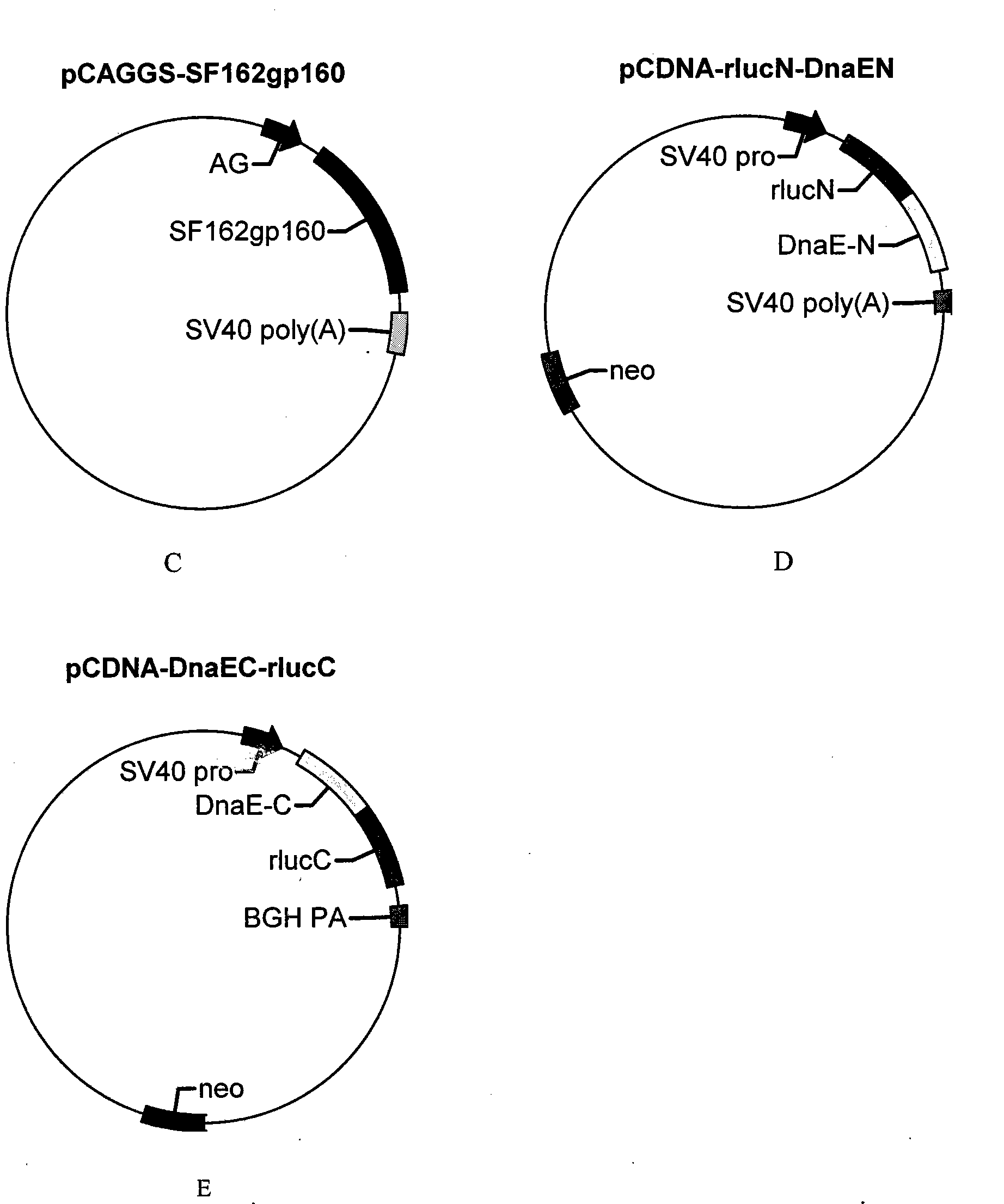
[0030] (2) Anacystis nidulans R2 dnae-C (1-36aa, see SEQ ID NO.9 for nucleotide sequence, see SEQ ID NO.10 for amino acid sequence) and Renilla luciferase gene C-terminal (rluc-C, 111-311aa , see SEQ ID NO.5 for the nucleotide sequence, and SEQ ID NO.6 for the amino acid sequence) splice dnae-C and rluc-C by overlapping PCR, and add 5 amino acids of CFNGT between dnae-C and rluc-C Sequence; the N-terminal of Renilla luciferase gene (rluc-N, 1-110aa, see SEQ ID NO.3 for nucleotide sequence, see SEQ ID NO.4 for amino acid sequence) and Anacystis nidulans R2 d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com