Method for quantitatively detecting chloramphenicol by utilizing photobacterium leiognathus
A quantitative detection and chloramphenicol technology, which is applied in the determination/inspection of microorganisms, chemiluminescence/bioluminescence, and analysis by making materials undergo chemical reactions, can solve the problems of environmental pollutant detection without luminescent bacteria
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
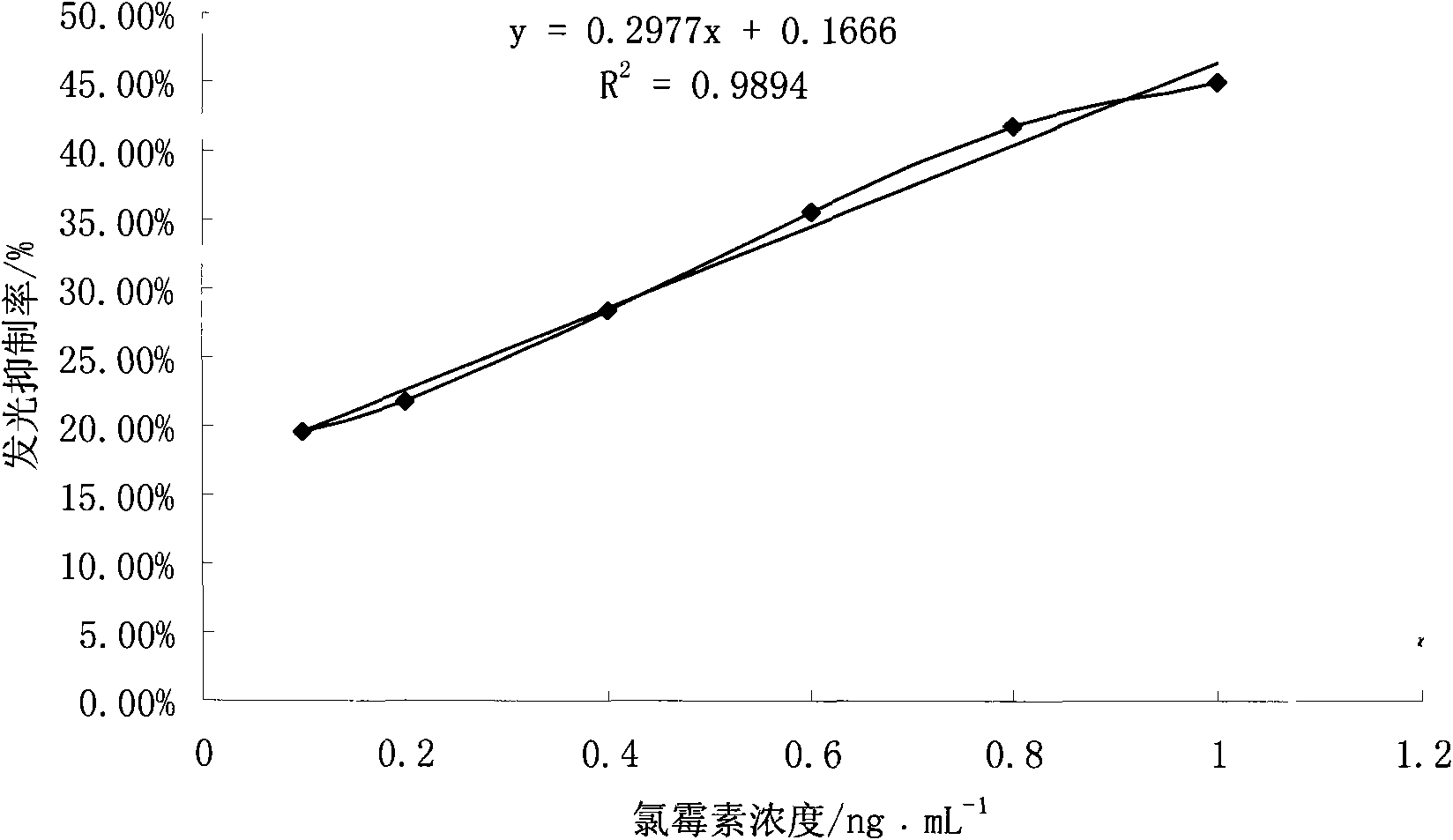
[0010] Inoculate P.leiognathi YL single colony in 150mL liquid medium (medium composition: peptone 5g, yeast extract 1g, FePO 4 0.1g, NaCl 20g, glycerol 3ml, distilled water 1000ml), 26°C, 150r / m shaker culture for about 6h, until the luminescence value reaches 2.0×10 5 -4.0×10 5 When the concentration of chloramphenicol is 0, 0.1ng / mL, 0.2ng / mL, 0.4ng / mL, 0.6ng / mL, 0.8ng / mL, 1.0ng / mL test tubes, the reaction system Chloramphenicol 5mL, bacteria solution 5mL. After mixing evenly, incubate at 26°C and 150r / m shaker for 30min, and measure the luminescence value. Draw the standard curve for chloramphenicol detection ( figure 1 )
Embodiment 2
[0012] Fish pretreatment method
[0013] (1) Take out the fish viscera, peel off the fish skin, and homogenize. Pack 20.0g of each bag of fish paste into small packaging bags for later use.
[0014] (2) Thaw the frozen fish, weigh 5.0g into a 50mL centrifuge tube, and add 50uL of standard CAP solution (concentration: 10ng / mL) to the centrifuge tube, which is equivalent to the concentration of chloramphenicol in the fish after adding the standard is 0.1ng / g, do three groups in parallel.
[0015] (3) Mix the samples in each centrifuge tube for 1 min.
[0016] (4) Add 20 mL of ethyl acetate to each centrifuge tube, mix for 1 min, sonicate for 10 min, and centrifuge for 5 min (4000 r / m).
[0017] (5) Pour the supernatant into a chicken heart bottle, and evaporate to dryness in a water bath at 40°C.
[0018] (6) Add 1 mL of methanol and vortex to dissolve the residue.
[0019] (7) Add 15 mL of n-hexane and mix evenly, then add 25 mL of 4% NaCl solution, shake and mix, and ext...
Embodiment 3
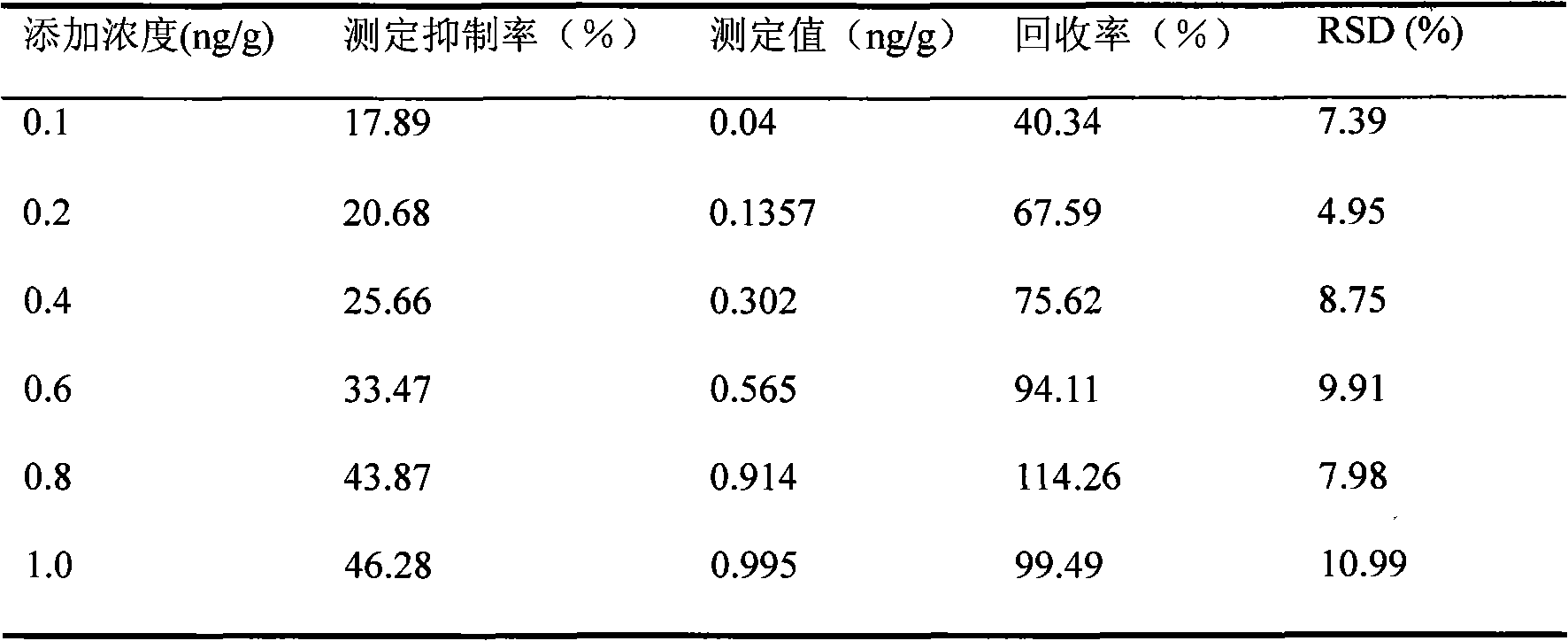
[0025] Thaw the frozen fish, weigh 5.0g into a 50mL centrifuge tube, add standard CAP solution (concentration: 10ng / mL) 50μL, 100μL, 200μL, 30μL, 400μL, 500μL to the centrifuge tube in turn, the The concentration of chloramphenicol was 0.1ng / g, 0.2ng / g, 0.4ng / g, 0.6ng / g, 0.8ng / g, 1.0ng / g, and six parallel groups were made for each concentration.
[0026] After adding chloramphenicol solution to the sample, pretreatment was carried out according to Example 2.
[0027] Inoculate a single colony of P.leiognathi YL into 150mL of liquid medium, put it into a shaking table for culture, and shaker parameter settings: 26°C, 150r / m. The luminous intensity of the P.leiognathi YL bacterial solution was at 3.0×10 5 Add 5mL of bacterial solution to each test tube containing 5mL of chloramphenicol solution recovered by standard addition, and make a set of standard solution blank control for each treatment method, that is, add 5mL of bacterial solution to a test tube containing 5mL of doubl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com