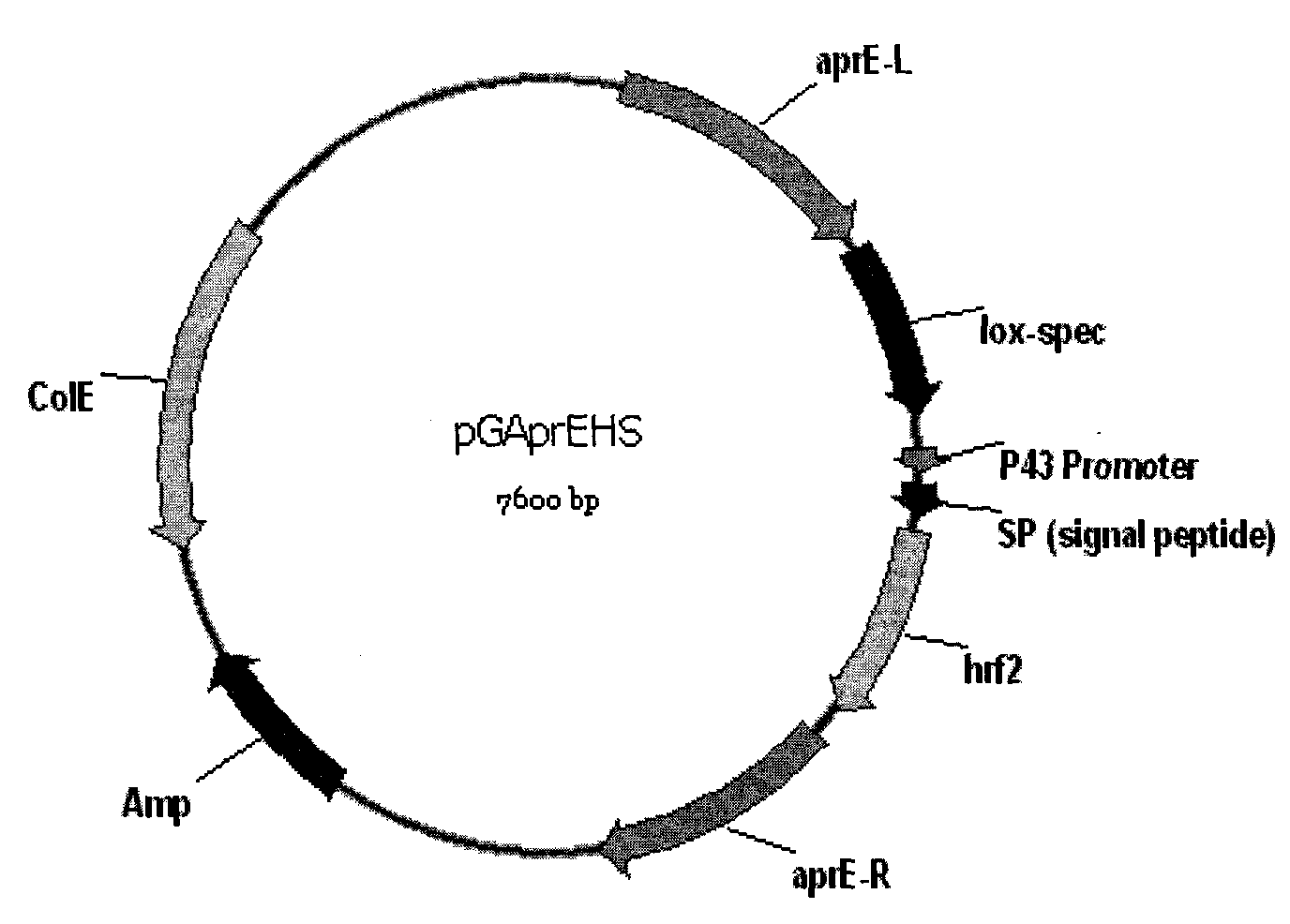
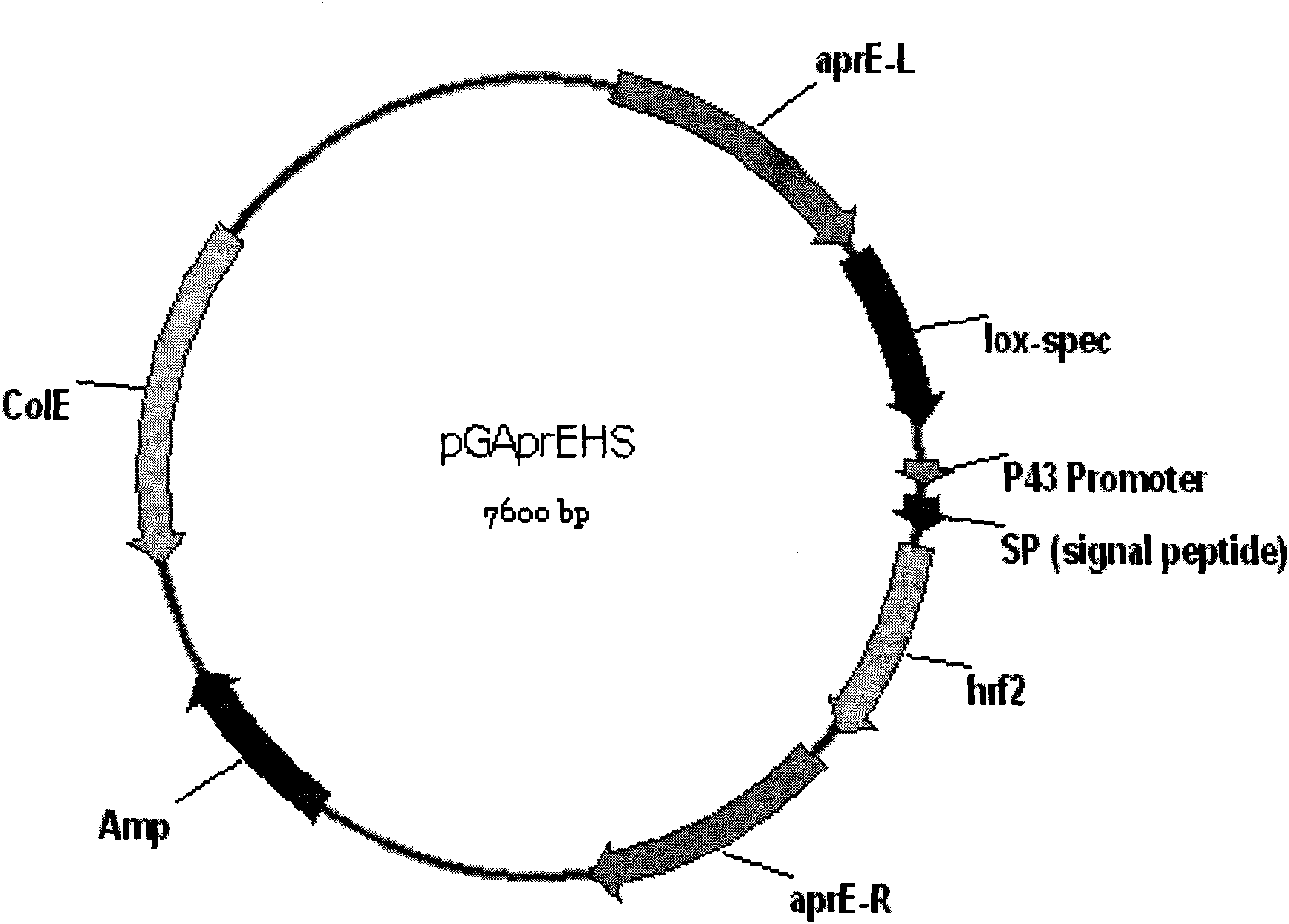
Recombinant vector pGAprEHS for expressing Harpin protein and engineering bacteria thereof
A technology of recombinant vectors and genetically engineered bacteria, applied in the field of genetic engineering, to achieve the effects of preventing and controlling plant diseases and insect pests, promoting plant growth, and increasing plant yield
Inactive Publication Date: 2010-08-25
无锡亚克生物科技有限公司
View PDF1 Cites 7 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
The research and development of Harpin protein is hailed as a green chemical revolution in plant production and food safety, and Bacillus subtilis is currently one of the only several microbial pesticides that can be used in the production of green vegetables A and AA, combining the two It is necessary to develop a green, environmentally friendly and multifunctional biological product, which can effectively promote plant growth, prevent and control plant diseases and insect pests, and increase plant yield when applied in fields and protected areas, but there are no relevant reports so far.
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment 1
Embodiment 2
Embodiment 3
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
The invention discloses a recombinant vector pGAprEHS for expressing Harpin protein and engineering bacteria thereof. An encoding gene of the Harpin protein is cloned into an expression vector so as to establish a bacillus subtilis recombinant vector pGAprEHS for expressing the Harpin protein; the bacillus subtilis recombinant vector pGAprEHS is then recombined into an alkaline proteolytic enzyme aprE gene locus on a bacillus subtilis chromosome to obtain intermediate engineering bacteria; an nprE gene locus mutation vector pUNprEK is then converted into the intermediate engineering bacteria so as to knock another intermediate proteolytic enzyme nprE gene out and establish bacillus subtilis gene engineering bacteria FZB42 / ANHSK which is deficient of two proteolytic enzymes and used for expressing the Harpin protein; and finally, an antibiotic screening mark is removed from the engineering bacteria by means of a Cre / lox recombinant enzyme system to obtain the engineering bacteria which are safer and more friendly to environment, people and livestock.
Description
technical field The invention belongs to the technical field of genetic engineering, and in particular relates to a recombinant vector pGAprEHS expressing Harpin protein and engineering bacteria thereof. Background technique Harpin protein is a protein encoded by some (or some) genes in the hrp gene cluster (hypersensitive reaction and pathogenicity gene cluster) in plant pathogenic bacteria. It is a non-specific elicitor and can induce plant disease resistance in non-host plants , can also initiate a variety of signal transduction pathways in plants, such as inducing plant production systems to acquire disease resistance (systemic acquired resistance, SAR) through the salicylic acid (salicylic acid, SA) signaling pathway, and promoting plant growth and Stress resistance, insect resistance is induced through jasmonic acid (jasmonic acid, JA) regulation signal pathway. In 1992, Dr. Zhongmin Wei of Cornell University in the United States first discovered that the hrpN gene p...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More IPC IPC(8): C12N15/75C12N1/21A01N63/00A01P21/00A01P3/00A01P1/00A01P7/04C12R1/125
Inventor 高学文乔俊卿伍辉军沈波
Owner 无锡亚克生物科技有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com