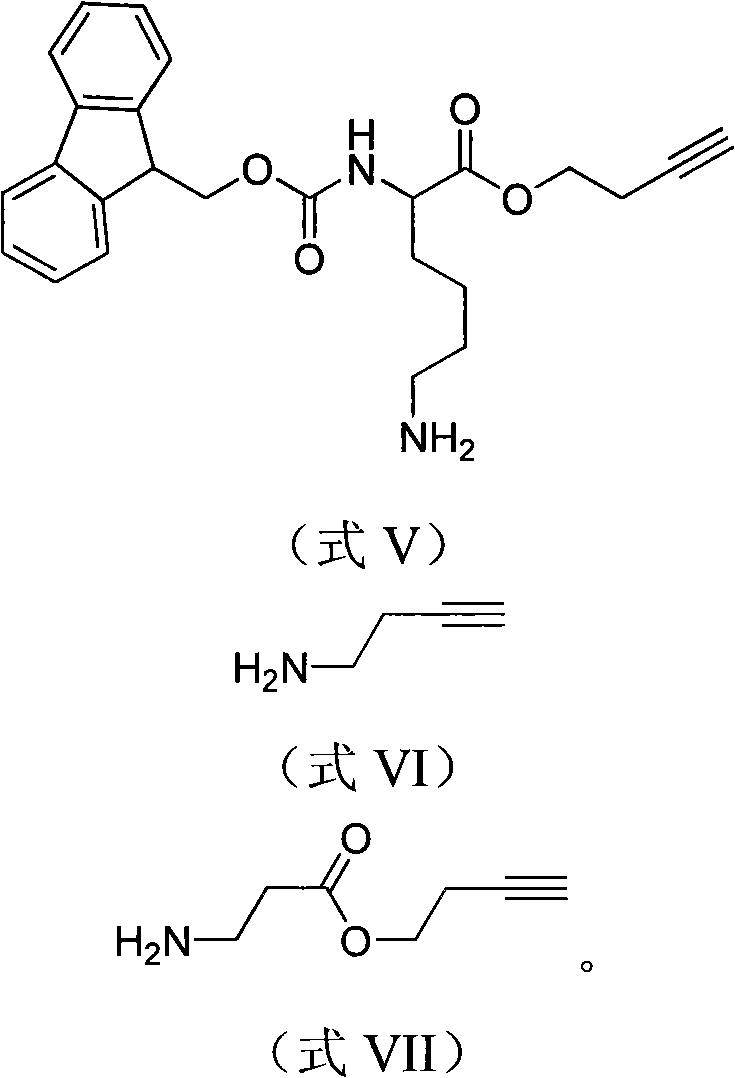
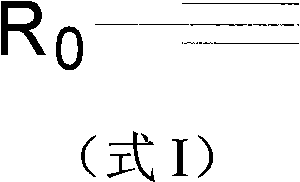Phosphorylated and/or glycosylated protein or peptide one-step enrichment modification determination method
A measurement method and glycosylation technology, which is applied in the field of site determination, can solve the problem that the hydrophilic properties of phosphopeptides are washed out, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
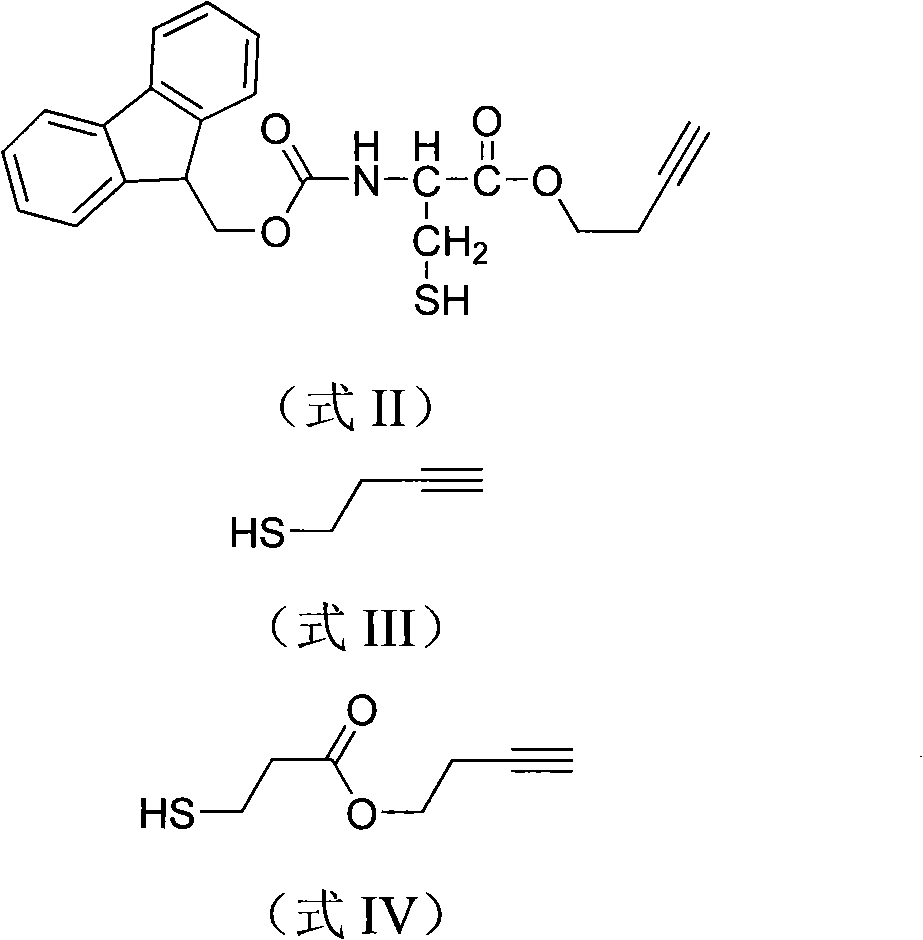
[0080] Embodiment 1, the terminal alkyne label compound shown in the synthetic formula II structural formula
[0081]
[0082] Add 585 mg of Fmoc-Cys(Trt)-OH (fluorenylmethoxycarbonyl cysteine mercaptotrityl) and 151 μL of 3-butyn-1-ol into the flask, then add dry acetonitrile, add 50 mg of DMAP (4 - dimethylaminopyridine). After the system was completely clarified, an appropriate amount of 2.2 g of DCC (1,3-dicyclohexylcarbodiimide) was added, and the temperature was naturally raised to room temperature. After reacting for 8 hours, the insoluble matter was removed by filtration, separated by a silica gel column, and the filtered solution was rotary evaporated with an oil pump, and the obtained product was drained by an oil pump, and weighed.
[0083] Separation was carried out on a silica gel column.
[0084] After the obtained product was dissolved in ethyl acetate, TFA (trifluoroacetic acid) was added and shaken for 2 h under the protection of argon. The material wa...
Embodiment 2
[0085] Embodiment 2, the terminal alkyne group label compound shown in the synthetic formula IV structural formula
[0086]
[0087] Add 210 μL of thioglycolic acid to 10 mL of 2N sodium hydroxide solution, slowly add 2.15 mL of p-toluenesulfonyl chloride dropwise while stirring in an ice-water bath, measure the pH value of the system after the dropwise addition, and add sodium hydroxide to control the pH value to 9 ~10. After stirring and reacting for 1 hour, wash off excess p-toluenesulfonyl chloride with ether, repeat the ether washing step twice, dilute with 15 mL ethyl acetate, acidify with 6M hydrochloric acid to pH 1, separate the organic phase and the aqueous phase Extract with ethyl acetate. The organic phases were combined, washed with 5% (wt) sodium chloride solution until neutral, dried over anhydrous sodium sulfate, and rotary evaporated to obtain a viscous liquid.
[0088] Add 10 mL of dry acetonitrile to the flask from the previous step, and add 50 mg of DM...
Embodiment 3
[0090] Example 3. Synthesis of the alkyne-terminated label compound SS-01 shown in the structural formula II
[0091]
[0092] 1. Methylbenzenesulfonyl protection of 3-butyn-1-ol
[0093] Add 4.55g of toluenesulfonyl chloride and 10mL of chloroform into a three-necked flask, stir well and cool to 3°C with an ice-water bath; drop 0.701g of 3-butyn-1-ol and 2.06g of pyridine into the flask, pass Ice-water bath and control the rate of addition to keep the reaction temperature at 6-8°C. After the dropwise addition, keep stirring at 8-10°C for 5 hours; pour the reaction mixture into 40mL ice water and wash it with stirring, then wash it twice with cold water, and adjust the pH value 8-9 sodium carbonate aqueous solution (temperature below 3°C) was fully stirred and washed twice, and finally washed with distilled water until the pH value was stabilized at 6.57-6.70; the water layer was separated by standing, and the organic layer was dried with anhydrous sodium sulfate. Atmosphe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com