MiRNA combination used for detecting esophageal squamous cell carcinoma and application thereof
A cell carcinoma, esophageal squamous cell technology, applied in recombinant DNA technology, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problems of tissue and cell detection defects, lack of human experience, improper material selection, etc., to achieve simple and easy effects. , easy to use, improve the accuracy of the effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
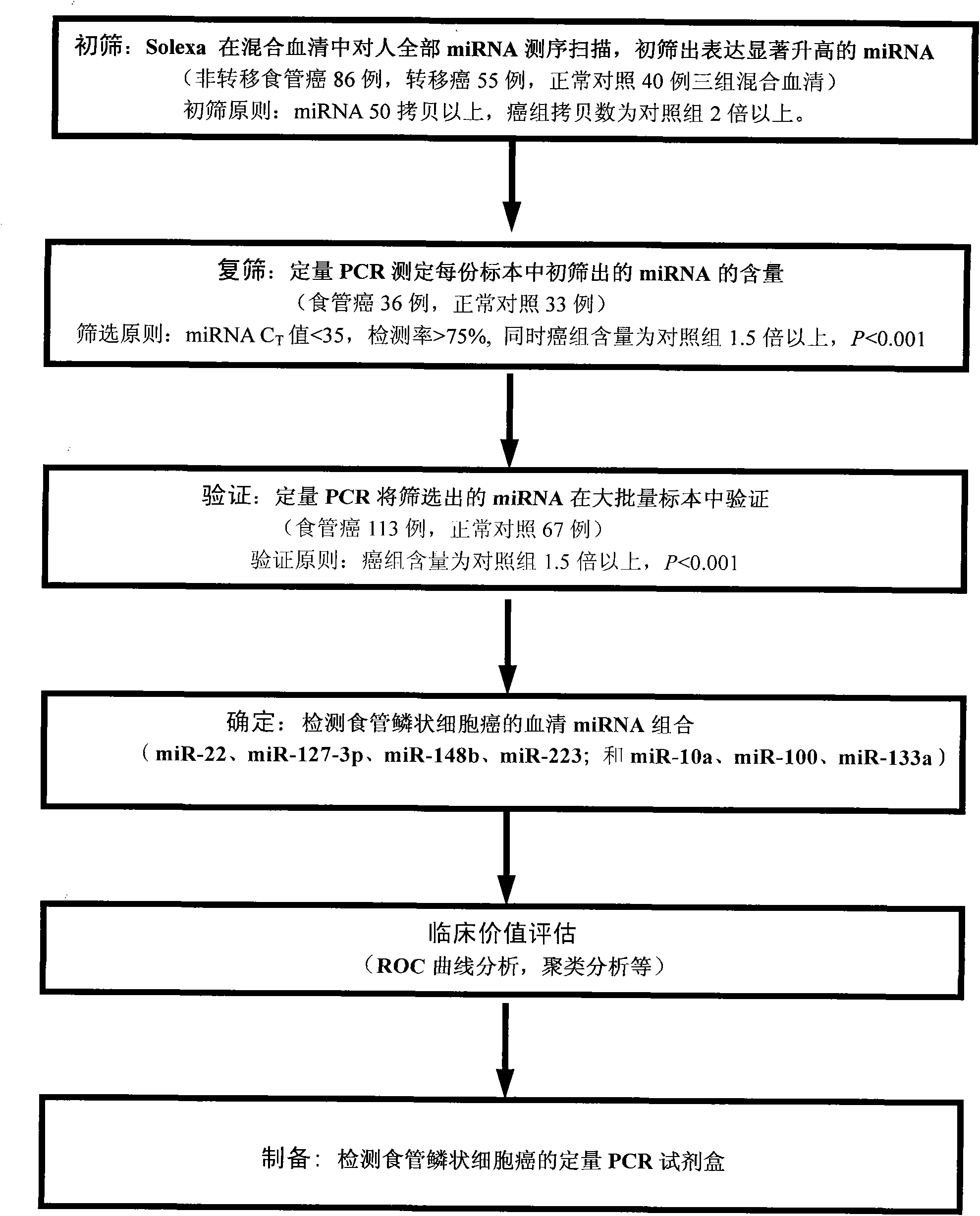
[0056] Example 1: Preliminary screening of specific miRNA expression profiles in esophageal squamous cell carcinoma
[0057] (1) 86 cases, 55 cases and 40 cases of serum samples of non-metastatic, metastatic esophageal squamous cell carcinoma and normal subjects were collected respectively, and the samples of the three groups were mixed respectively;
[0058] (2) Extract the RNA in the mixed serum of the three groups respectively. The specific plan is: use Trizol reagent (Invitrogen Company) to extract the total RNA, and use the phenol-chloroform three-step method to further remove impurities such as protein and purify the RNA from the supernatant;
[0059] (3) Solexa sequencing analysis was performed on the total RNA of the mixed serum of the three groups.
[0060] The specific plan is: Solexa sequencing is based on single-molecule cloning array technology, which can distinguish the difference of a single base at the level of the entire genome, thereby distinguishing the diff...
Embodiment 2
[0067] Embodiment 2: Real-time fluorescent quantitative PCR experiment (TaqMan probe method) of miRNA in serum
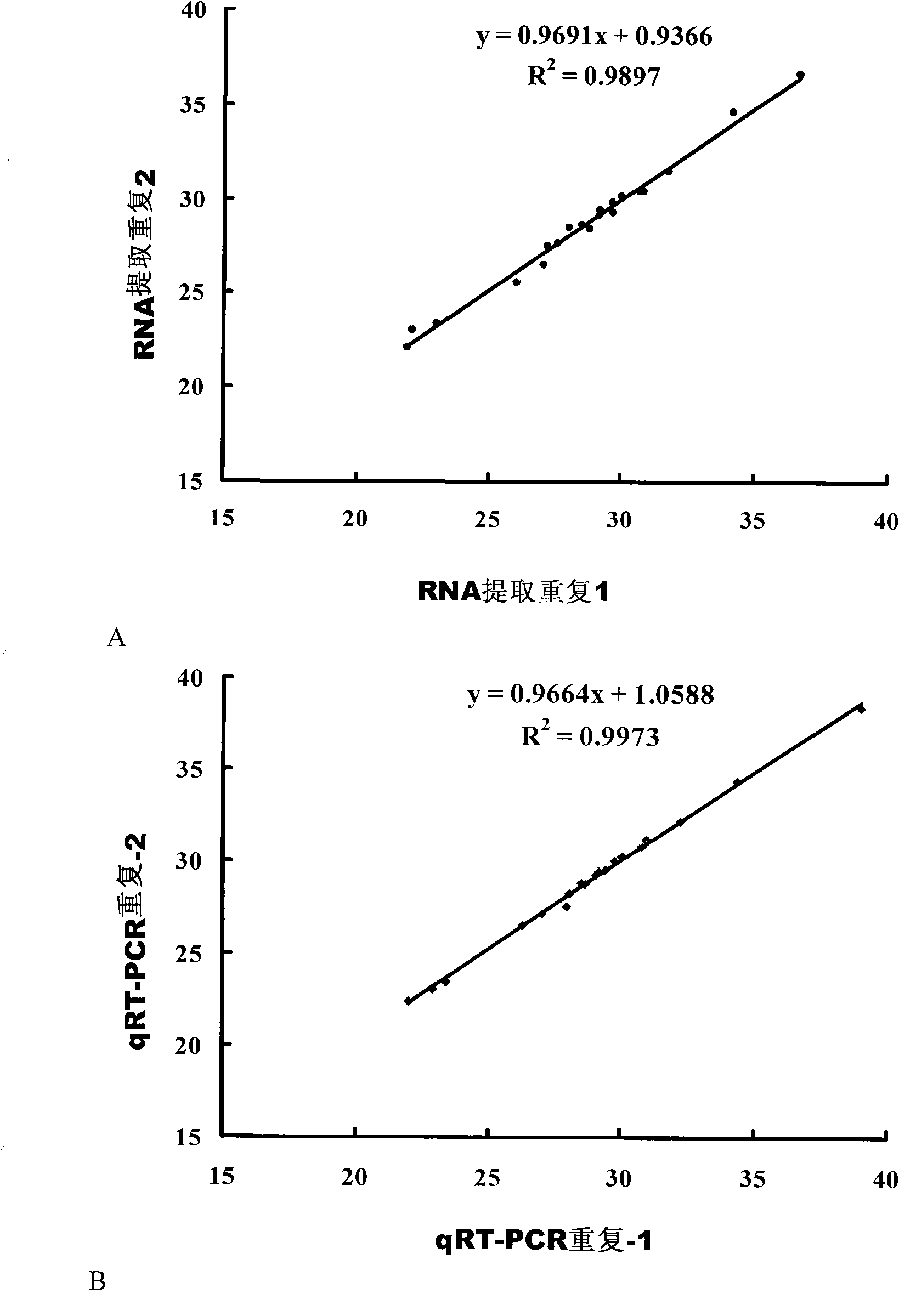
[0068] Quantitative PCR was used to verify batches of samples one by one, and miRNAs with significant expression differences were screened from the miRNAs initially screened by the Solexa method.
[0069] Firstly, the extracted serum total RNA was reverse transcribed into cDNA. For each miRNA, design a gene-specific reverse primer containing the same stem-loop structure, use the miRNA-specific reverse primer to perform reverse transcription, and obtain a cDNA (product of ABI Company) that contains a common stem-loop structure but belongs to a specific miRNA.
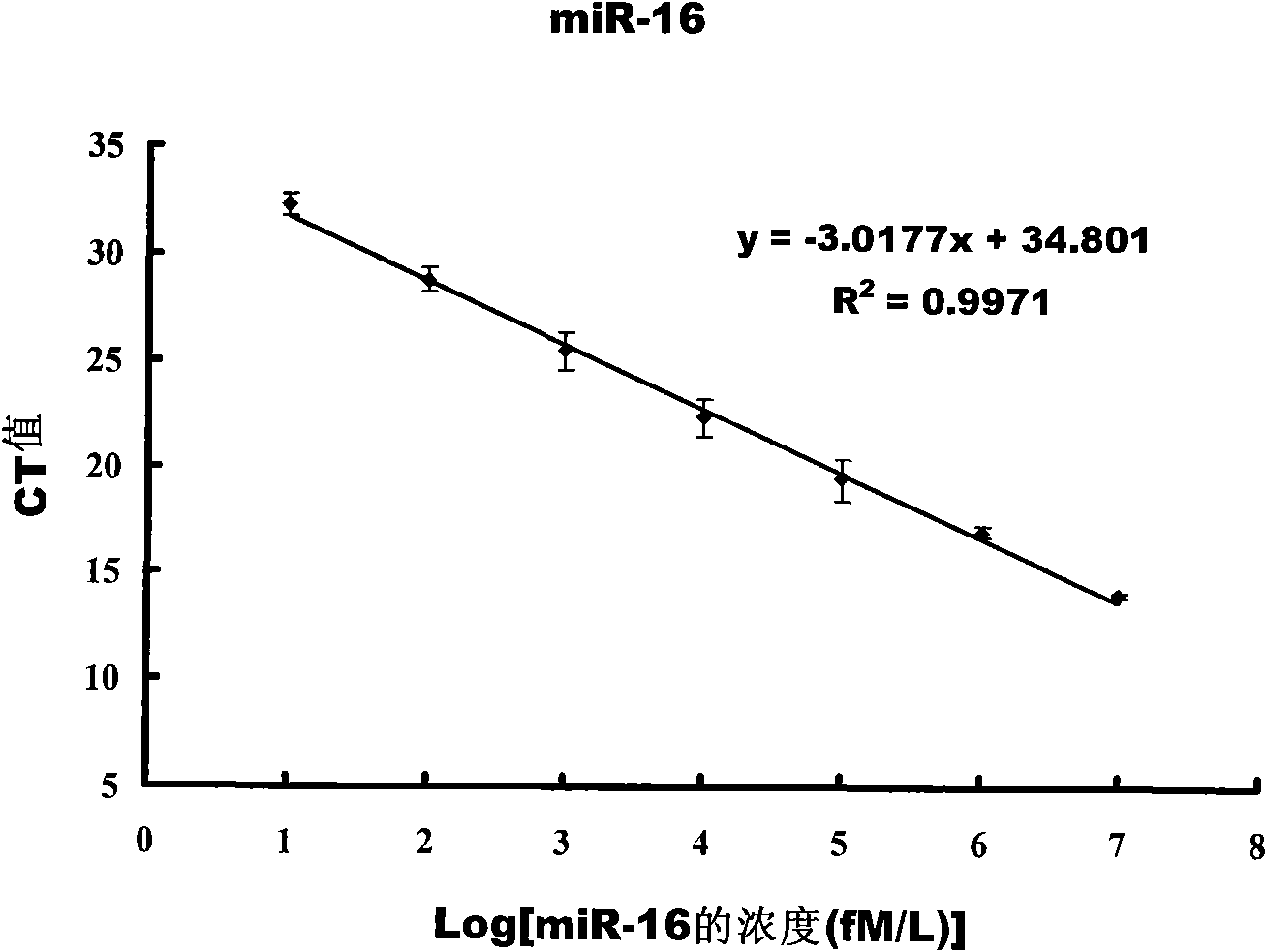
[0070] The instrument used is ABI Prism 7300 fluorescent quantitative PCR instrument.
[0071] In this experiment, different concentrations of artificially synthesized mature miR-16 were used as a standard curve. The artificially synthesized miR-16 stock solution was reverse-transcribed into cDNA, and dilute...
Embodiment 3
[0075] Embodiment 3: Quantitative PCR method re-screens the serum miRNA screened out
[0076] A group of subject samples were detected one by one by TaqMan quantitative PCR method, and miRNAs with significant expression differences between the patient group and the control group were screened from the 25 miRNAs initially screened by the Solexa method (see Example 2 for specific steps). There were 36 patients with esophageal squamous cell carcinoma in this group, and 33 cancer-free patients matched in age and gender were used as controls (see Table 2 for detailed clinical information of patients). It was found that among the 25 miRNAs initially screened out, the expression levels of 7 miRNAs in the patient group were significantly higher than those in the control group (the expression method of content is mean ± standard deviation; the multiple of change is 1.65-2.97 times, p<0.0001). The seven miRNAs are miR-10a, miR-22, miR-100, miR-148b, miR-223, miR-133a and miR-127-3p (see...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com