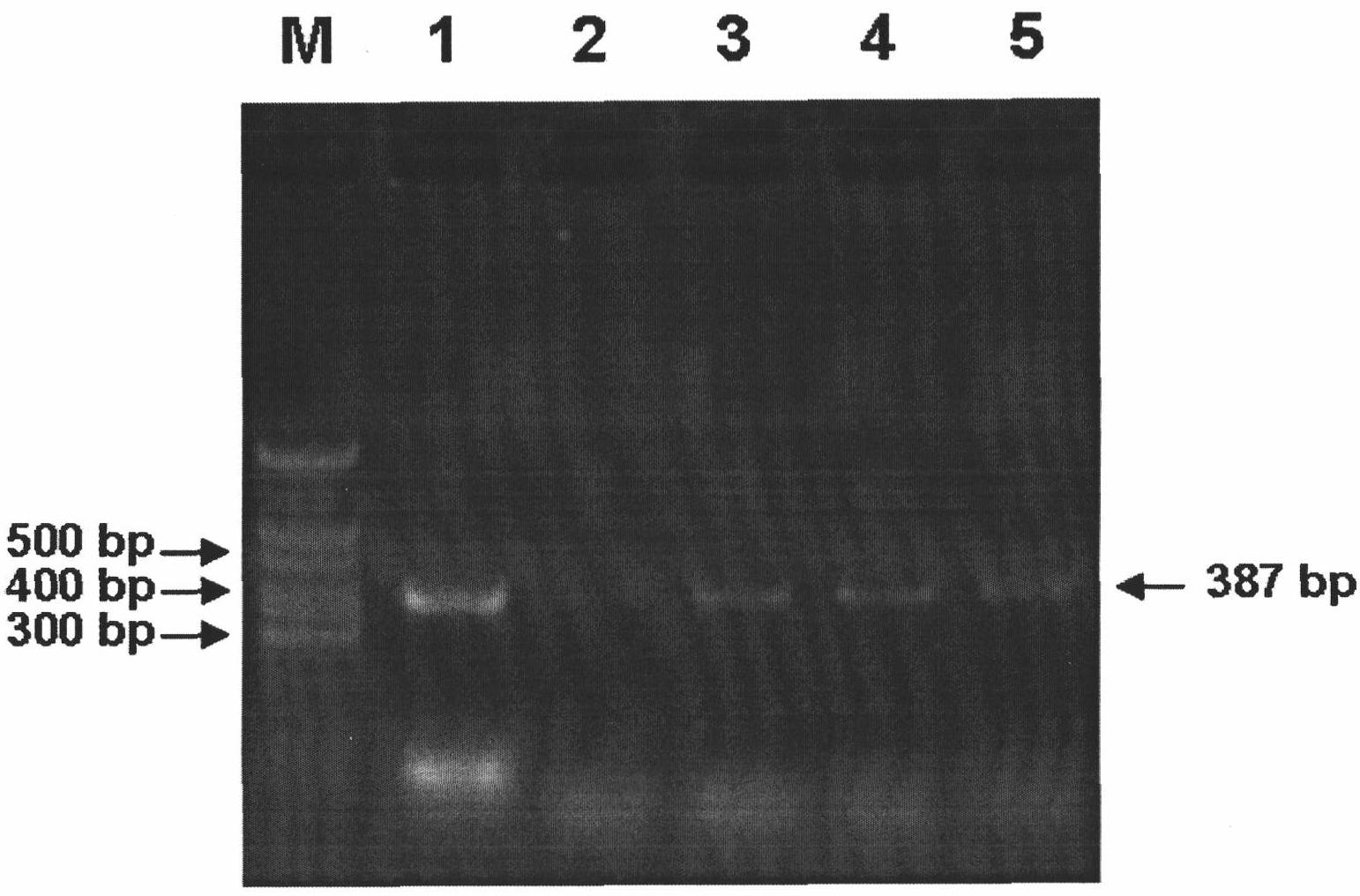
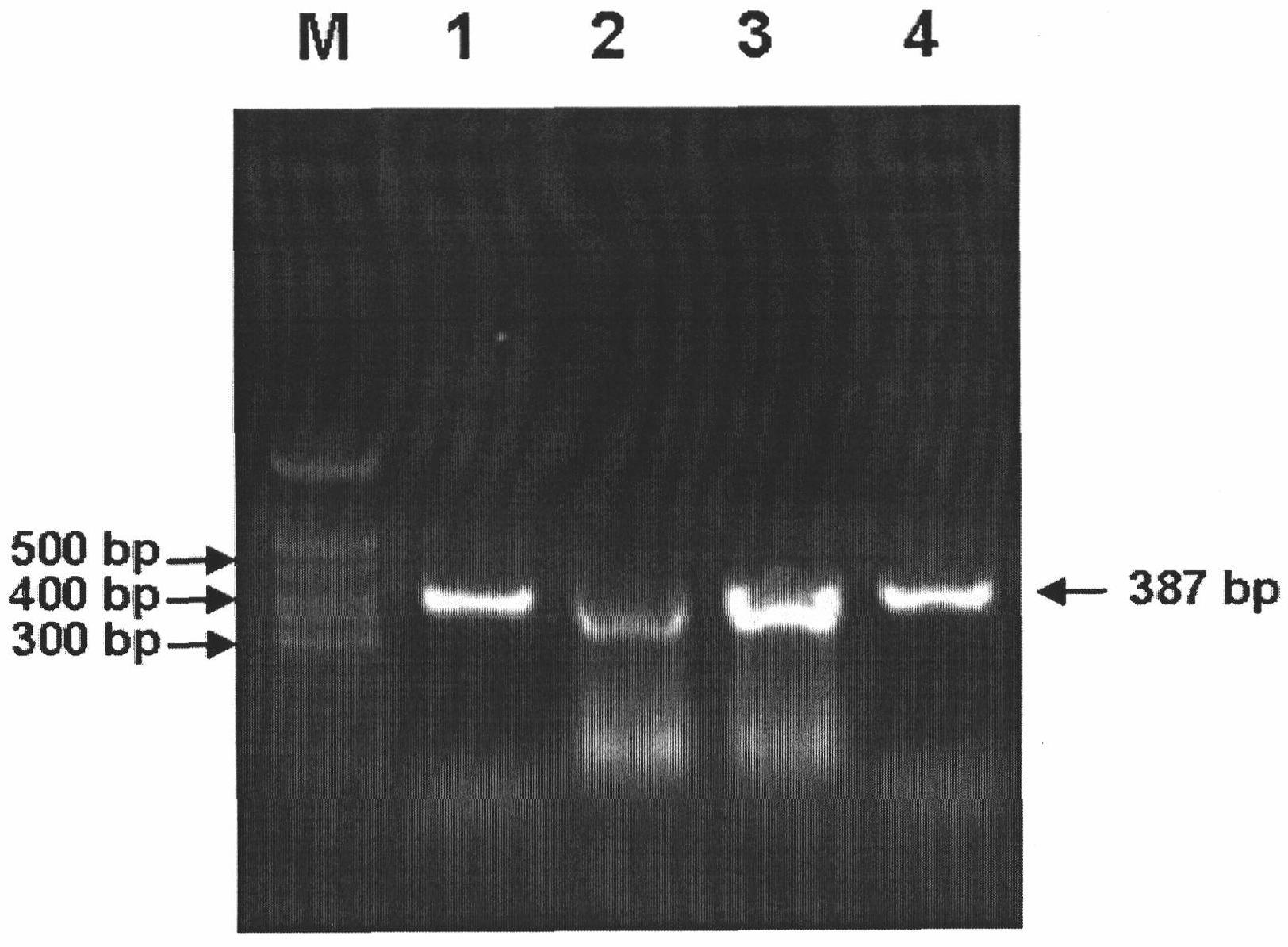
Method for detecting avian paramyxoviruses
An avian paramyxovirus and detection kit technology, which is applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc. , high sensitivity and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
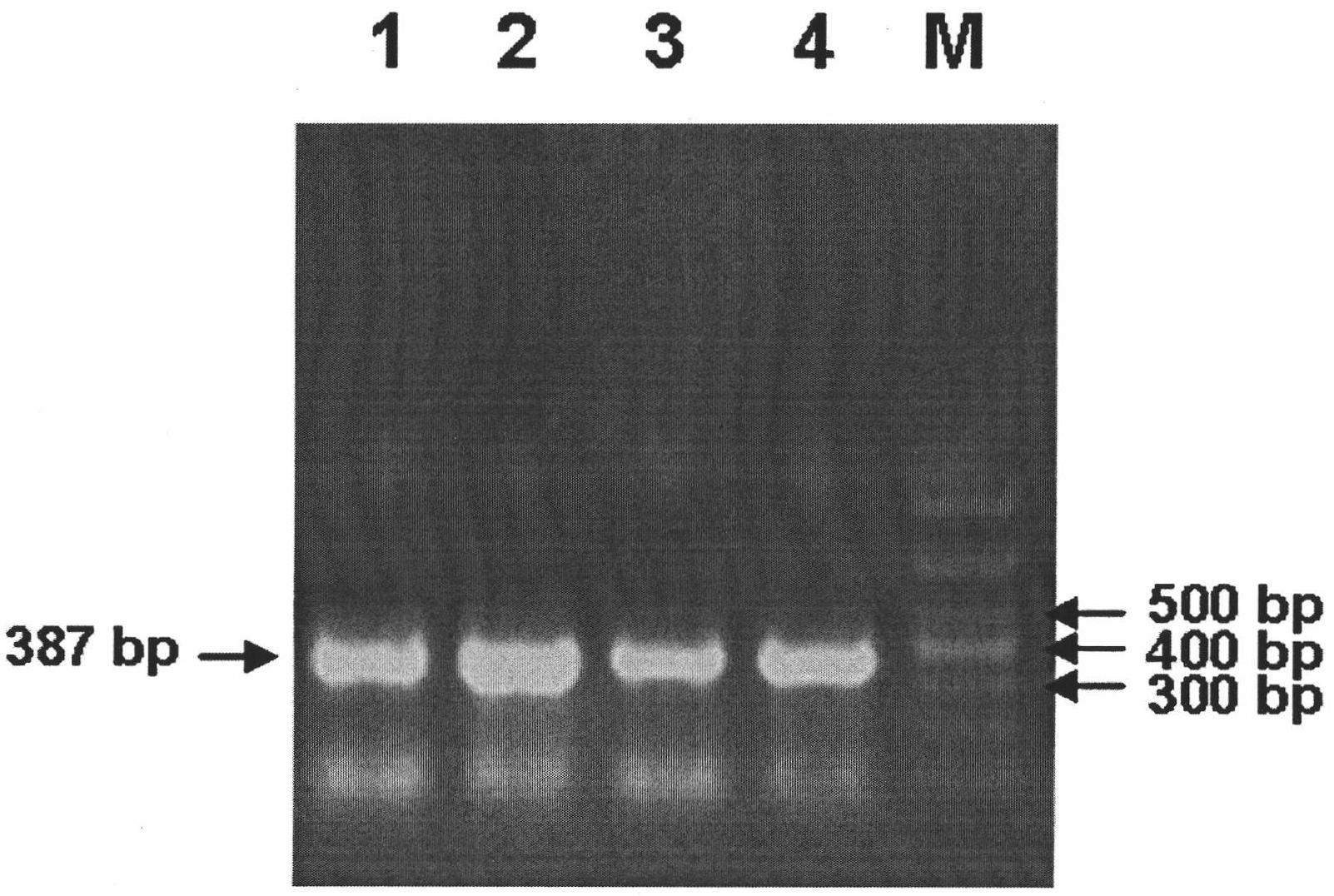
Image
Examples
Embodiment 1
[0018] The pretreatment of embodiment 1 sample
[0019] 1. Experimental reagents and main instruments
[0020] Main reagents: sterile saline
[0021] Main instruments: Centrifuge 5810R low-temperature desktop centrifuge, a product of Eppendorf, Germany; vortex oscillator; grinder
[0022] 2. Experimental steps
[0023] (1) Tissue sample processing: take 1 mg of organ tissue sample, add 1 ml of sterilized saline, grind and suspend with a grinder, centrifuge the tissue suspension at 3000 rpm for 30 min, and take the supernatant for detection.
[0024] (2) Cloacal or oropharyngeal swab sample processing: add 0.5ml sterilized physiological saline to the swab sample and shake and suspend it with a vortex shaker. The sample suspension is centrifuged at 3000rpm for 30min and the supernatant is taken for detection.
Embodiment 2
[0025] Example 2 Extraction of sample total RNA
[0026] 1. Experimental reagents and main instruments
[0027] Main reagents: Trizol RNA extraction reagent, Invitrogen product, purchased from Beijing Tianyouda Company; DEPC treated water, purchased from Beijing Santai Xinglong Technology Co., Ltd.; new chloroform, isopropanol and absolute ethanol; 75% ethanol; Rnase- free centrifuge tubes and tips
[0028] Main instruments: Centrifuge 5810R low-temperature desktop centrifuge, product of Eppendorf, Germany; biological safety cabinet, product of Forma Scientific
[0029] 2. Experimental steps
[0030] (1) Take 250 μl of the suspension of the sample to be tested, add 750 μl Trizol, invert and mix for 15 seconds until the liquid becomes viscous, and ice-bath for 15 minutes;
[0031] (2) Add 200 μl chloroform, mix by inversion for 15 seconds, and ice bath for 15 minutes;
[0032] (3) The mixture was separated into two phases by centrifugation at 12000 rpm and 4°C for 15 minutes;...
Embodiment 3
[0039] Example 3 Reverse transcription to generate cDNA
[0040] 1. Experimental reagents and main instruments
[0041] Main reagents: reverse transcriptase (Reverse Transcriptase) 200U / μl (Promega); nuclease inhibitor (Rnase Inhibitor) 50U / μl (Takara); 5 times the volume of reaction buffer (5×Reaction Buffer); dNTP mixture 2.5 mM, purchased from Beijing Qiangxin Borui Biotechnology Co., Ltd.; Random Primer (Random Primer) 500 μg / ml (Promega), which is a random hexamer; DEPC-treated water, purchased from Beijing Santai Xinglong Technology Co., Ltd.
[0042] Main instruments: PCR amplification instrument (T-Gradient Thermoblock), purchased from Beijing North Huaao Trading Co., Ltd.; small desktop centrifuge (Eppendorf, Germany); DK-8D electric heating constant temperature water tank (Shanghai Senxin Experimental Instrument Co., Ltd. ).
[0043] 2. Experimental steps
[0044] (1) Add the following ingredients into a 0.2ml centrifuge tube:
[0045] RNA solution 6μl
[0046] ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Upstream primer | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com