Kit for detecting 7 hot mutant sites of phenylketonuria PAH gene and PCR amplification method thereof
A technology for phenylketonuria and mutation sites, applied in the field of clinical detection technology, can solve the problems of difficult popularization and application, time-consuming, low efficiency, etc., and achieve the effects of high throughput, simple operation and high specificity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
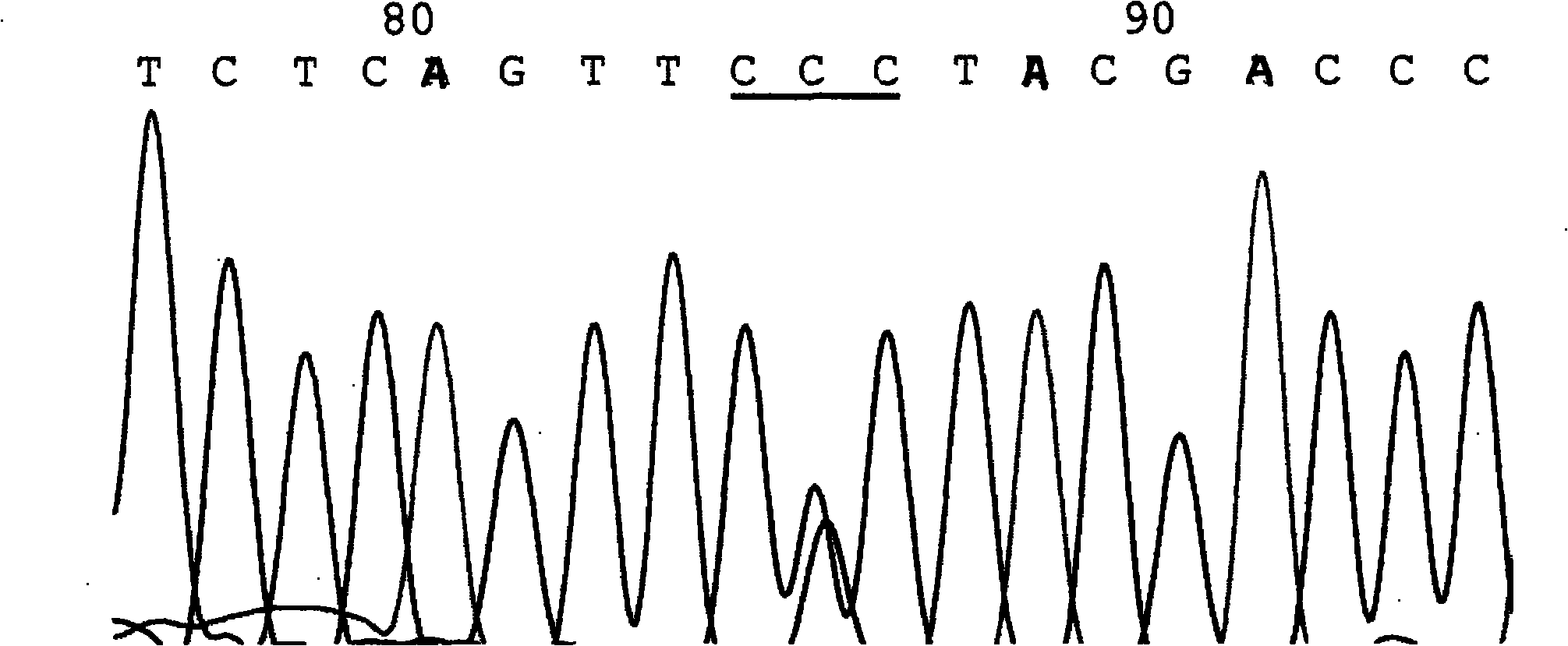
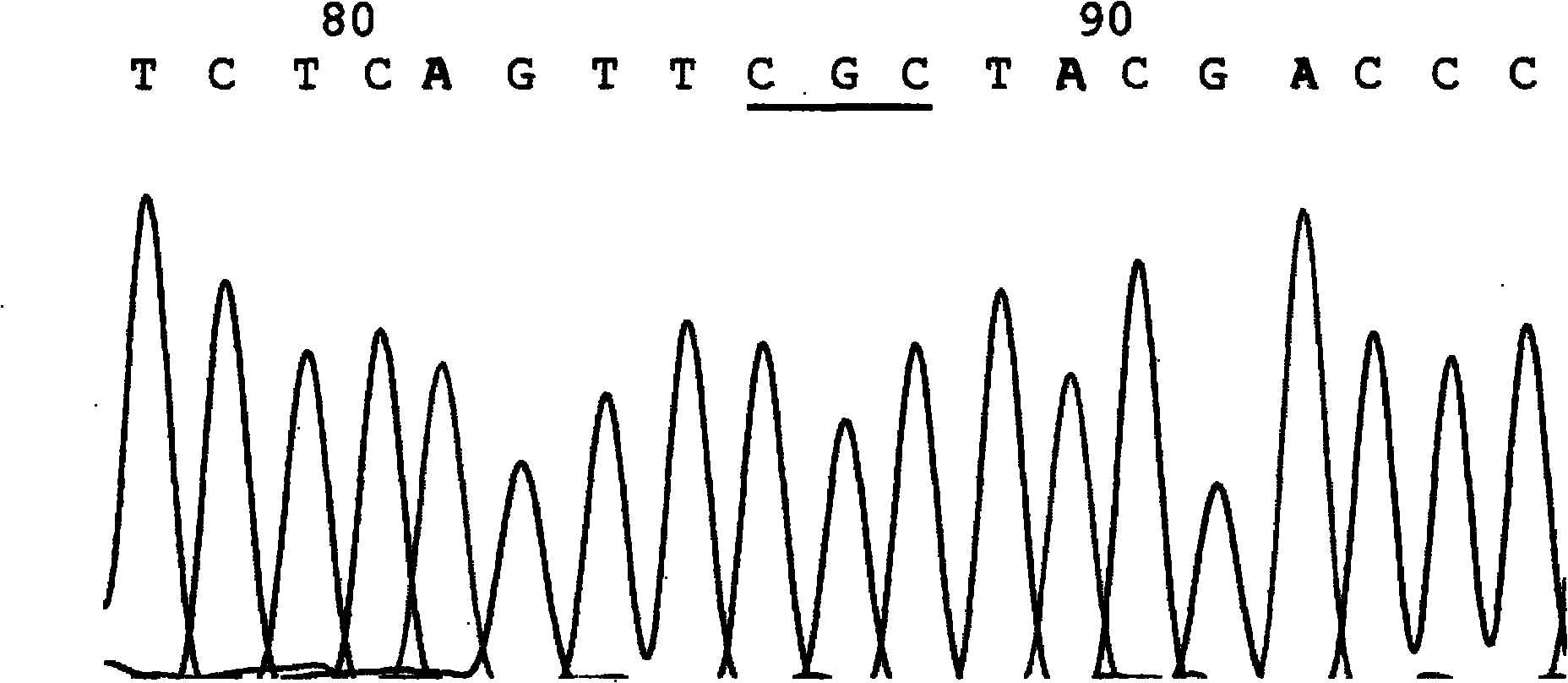
[0103] Embodiment one: a kind of test kit that detects seven hotspot mutation sites of phenylketonuria PAH gene, described kit: by 10 * PCR Buffer, dNTP (each 2.5mM), MgSO 4 (25mM), seven sets of primers (10mM each) and Pfu DNA polymerase (5U / μl).
[0104] The seven sets of primers refer to seven sets of primers for detecting seven hotspot mutation sites of R111X, IVS-4-1, Y204C, R243Q, W326X, Y356X and R413P;
[0105] The first group: the nucleotide sequence of the primers for detecting the R111X hotspot mutation site is as follows:
[0106] Mutation template paired primer R111X M:
[0107] 5'-ACCTGTGTCTTTCTTCTTATCTC*AA-3';
[0108] Normal template pairing primer R111X N:
[0109] 5'-ACCTGTGTCTTTCTTCTTATCTC*GA-3';
[0110] Common upstream or downstream primer R111X C:
[0111] 5'-TGCCCCACCTCCTGCCACTT-3';
[0112] Wherein, the phosphodiester bond between the -3 and -2 bases at the 3' end of the mutant template matching primer R111X M and the normal template matching prim...
Embodiment 2
[0158] Embodiment two: a kind of PCR amplification method that adopts the kit described in embodiment one to detect phenylketonuria PAH gene hotspot mutation site, comprises the following steps:
[0159] Step 1: Prepare DNA
[0160] (1) Blood is drawn from two human bodies to obtain two blood samples.
[0161] (2) Obtaining DNA from two blood samples, that is, preparation of genomic DNA samples of white blood cells in the blood samples.
[0162] Reagent preparation:
[0163] Anticoagulant: Each 100ml anticoagulant contains 0.48g citric acid, 1.32g sodium citrate, and 1.47g glucose.
[0164] Red blood cell lysate: 10mmol / L Tris-HCl, pH 7.6;
[0165] 5mmol / L MgCl 2 ;
[0166] 10mmol / L NaCl;
[0167] White blood cell lysate: 10mmol / L Tris-HCl, pH 7.6;
[0168] 10mmol / L EDTA (pH 8.0)
[0169] 50mmol / L NaCl
[0170] 10mg / ml proteinase K (Protease K): 10mg Protease K dissolved in 1ml ddH 2 O (double distilled water), aliquoted and stored at -20°C. When in use, melt at 4°C. ...
Embodiment 3
[0203] Embodiment 3: A PCR amplification method for detecting hotspot mutation sites of PAH gene in phenylketonuria using the kit described in Embodiment 1
[0204] The experimental conditions and judgment methods are the same as in Example 2, and there are two blood samples (different from Example 2).
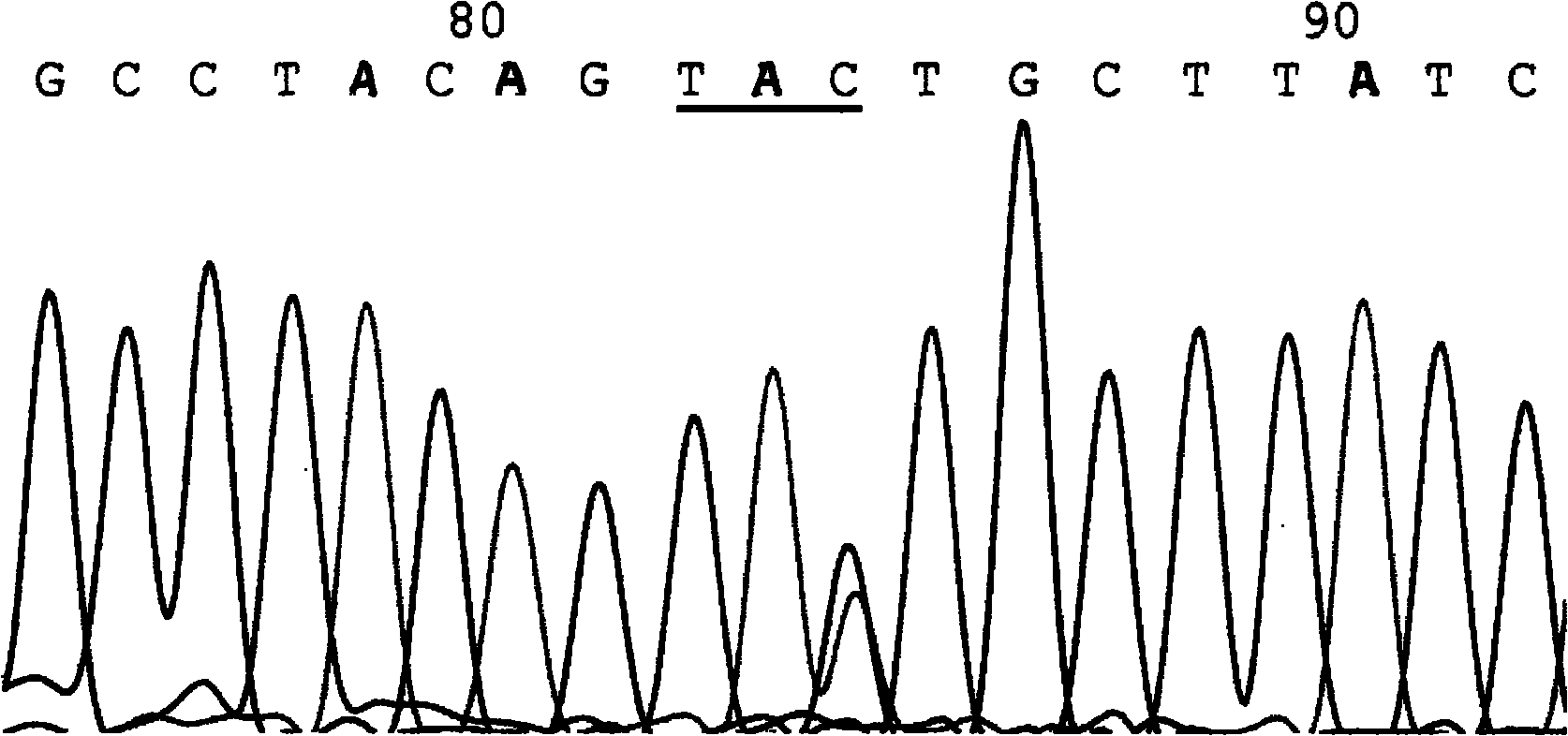
[0205] The following set (two pairs) of primers were used to detect the mutation site of IVS-4-1 (AG→AA). The size of the product fragment corresponding to each pair of primers was 176bp, and its nucleotide sequence was (5’→3’):
[0206] ①. Mutation template matching primer pair:
[0207] Mutation template paired primer IVS-4-1M: CAGGTGTCTCTTTTCTCCTA*AC
[0208] Common upstream or downstream primer IVS-4-1C: TTCCATCCTCAACTGGATGA
[0209] ②, normal template matching primer pair:
[0210] Normal template paired primer IVS-4-1N: CAGGTGTCTCTTTTTCTCCTA*GC
[0211] Common upstream or downstream primer IVS-4-1C: TTCCATCCTCAACTGGATGA
[0212] Note: The phosphodiester bond marked ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com