Heparanase II fusion protein and coding gene and expression method thereof
A technology of heparinase and protein, applied in the fields of genetic engineering and fermentation engineering, can solve the problems of limited research on heterologous recombinant expression of heparinase II, low expression amount and high purification cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
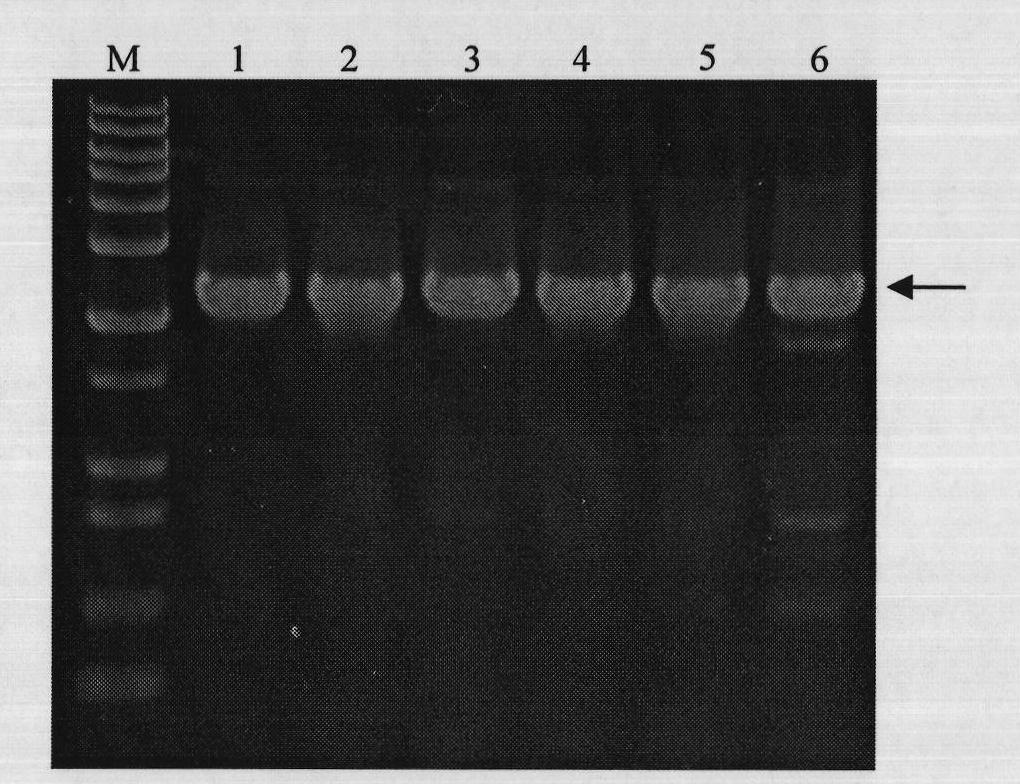
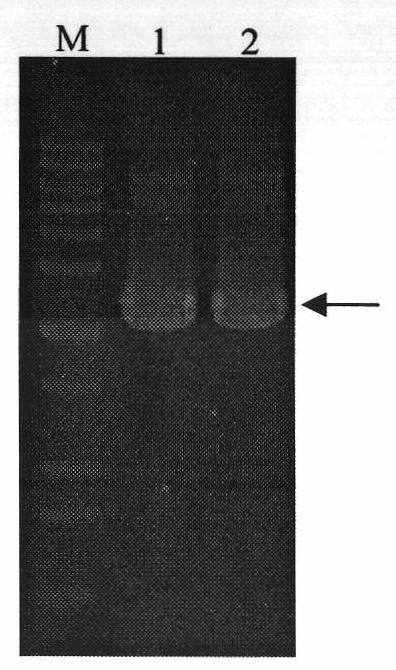
[0039] Embodiment 1, expression of heparanase II fusion protein MBP-HepB
[0040] 1. Cloning of Flavobacterium heparinase II coding sequence with signal peptide removed
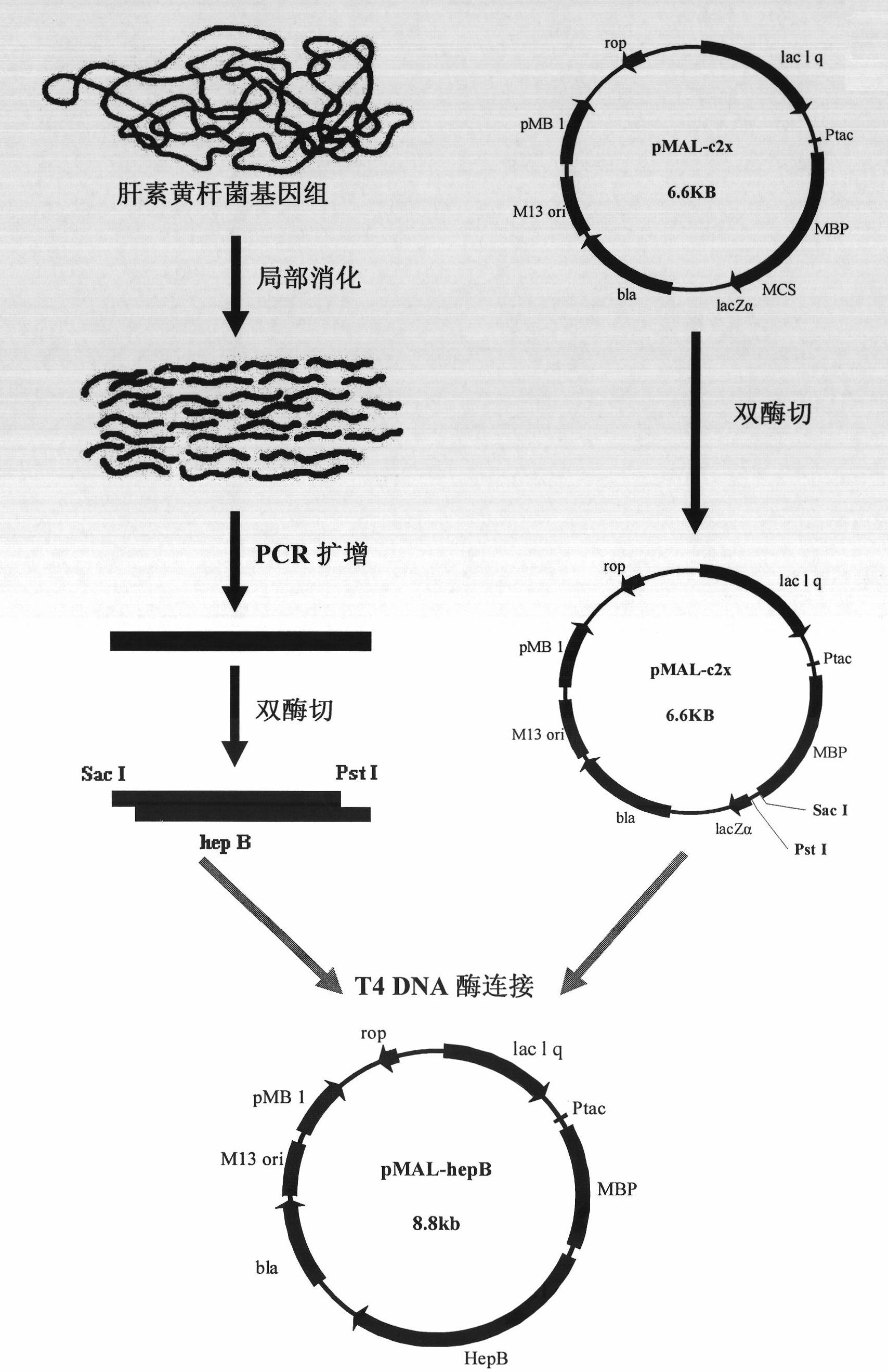
[0041] The construction process of the expression vector pMAL-hepB is as follows: figure 1 As shown, the specific process is as follows:
[0042] 1. Design and synthesis of primers
[0043] The DNA sequence of heparinase II from Flavobacterium heparinum (Su, H., Blain, F., Musil, R.A., Zimmermann, J.J., Gu, K. and Bennett, D.C. coli of hepB and hepC, genes coding for the glycosaminoglycan-degrading enzymes heparinase II and heparinase III, respectively, from Flavobacterium heparinum.Appl.Environ.Microbiol.1996, 62, 2723-2734), and then according to the removal of the coding signal peptide base Design primers for the DNA sequence of Flavobacterium heparinase II heparinase, and introduce the recognition sites of restriction endonucleases Sac I and Pst I in the primer sequences, and the upstream and downstrea...
Embodiment 2
[0061] Example 2. Purification of heparanase II fusion protein MBP-HepB by amylose column
[0062] The fusion partner (fusion partner) maltose binding protein MBP utilized in the present invention can achieve one-step separation with amylose affinity adsorption. The specific steps of affinity separation are as follows: 100 mL of bacterial cells with a final concentration of 0.24 mM IPTG induced expression for 24 hours were centrifuged at 10000 rpm for 5 minutes; at the same time, a bacterial cell without induced expression was set as a control. Then proceed with the following two options:
[0063] Option 1: Wash twice with Column buffer (20mM Tris-HCl, 200mM NaCl, pH7.5), resuspend in 5mL Column buffer, and perform sonication (300W output power, 3 seconds each time and intermittent 198 times in 3 seconds).
[0064] Option 2: Osmotic pressure shock. Resuspend the bacteria in 100 mL osmotic shock buffer I (20-40% sucrose, 30 mM Tris-HCl, 1 mM EDTA) for 15 minutes, and stir. ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com