Genetic detection method of powdery mildew-resistant near-isogenic lines Brock/Jing411<7>
A powdery mildew resistance gene and base sequence technology, which is applied in the field of molecular detection and application of genetic background of gene lines, can solve the problems of long cycle, low selection efficiency, developmental and environmental conditions, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment 1, the preparation of near isogenic line:
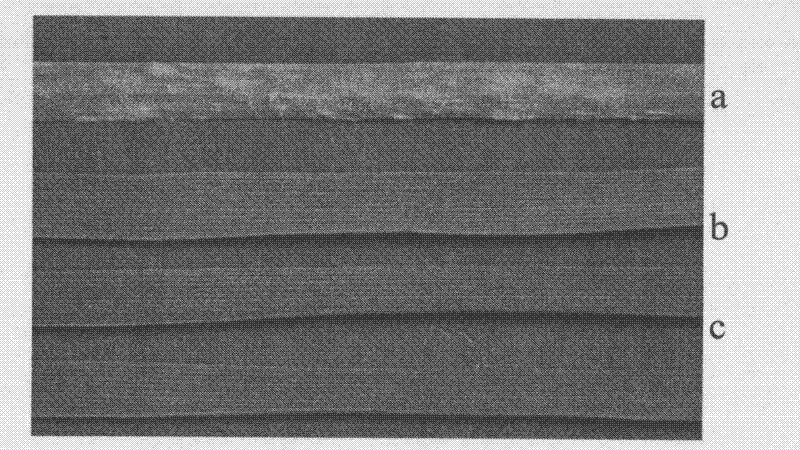
[0037] The preparation of the near-isogenic line of the present invention: the parent of the disease-resistant gene donor is from the British common wheat variety Brock, and it is crossed with the parent Jing 411, which has excellent agronomic traits but is susceptible to the disease, and the hybrid combination is "Brock / Jing 411", and the obtained F 1 They are all resistant to powdery mildew. Backcross with Beijing 411, under the stress of Physiological race No. 15 of powdery mildew that is prevalent in North China, select a single disease-resistant plant, and after 7 generations, self-cross again to obtain the powdery mildew resistant near-isogenic line Brock / Beijing 411 7 . figure 1 It is Jing 411, near-isogenic line Brock / 015 / / Jing 411 7 , near isogenic line Brock / Jing 411 7 Compared with Brock leaves infected with bacteria for 2 weeks, it was found that 411 was seriously infected with bacteria, and the hyp...
Embodiment 2
[0038] Embodiment 2, the separation of genomic DNA:
[0039] Take 0.2g of etiolated seedling leaves and grind them in liquid nitrogen until they are as powder as possible, quickly add 2ml of extraction buffer (50mM Tris-HCl, pH 8.0; 20mM EDTA; 50mM NaCl), mix gently, and incubate at 65°C for 15min, during which Mix by inversion 2-3 times. Ice bath at 0°C for 10 minutes, centrifuge at 0-4°C at 3000 rpm for 15 minutes, remove the supernatant, add it to a new tube, add an equal volume of Tris-HCl (pH 8.0) saturated phenol, and mix gently. Centrifuge at 0-4°C at 3000 rpm for 10 minutes, take the supernatant, add an equal volume of phenol, chloroform-isoamyl alcohol (25:24:1) for extraction, and mix gently. Centrifuge at 0-4°C at 3000 rpm for 10 minutes, take the supernatant, add an equal volume of chloroform-isoamyl alcohol (25:24:1) for extraction, and mix gently. 0-4°C, 3000 rpm, centrifuge for 10 minutes, take the supernatant, add 2 times the volume of cold ethanol, and preci...
Embodiment 3
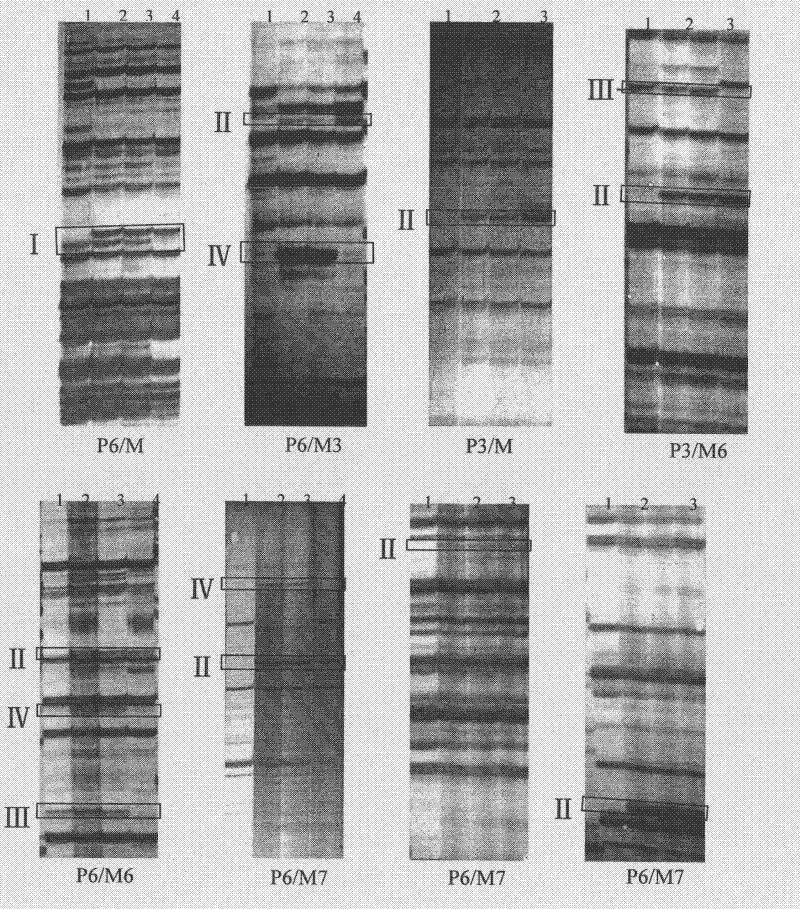
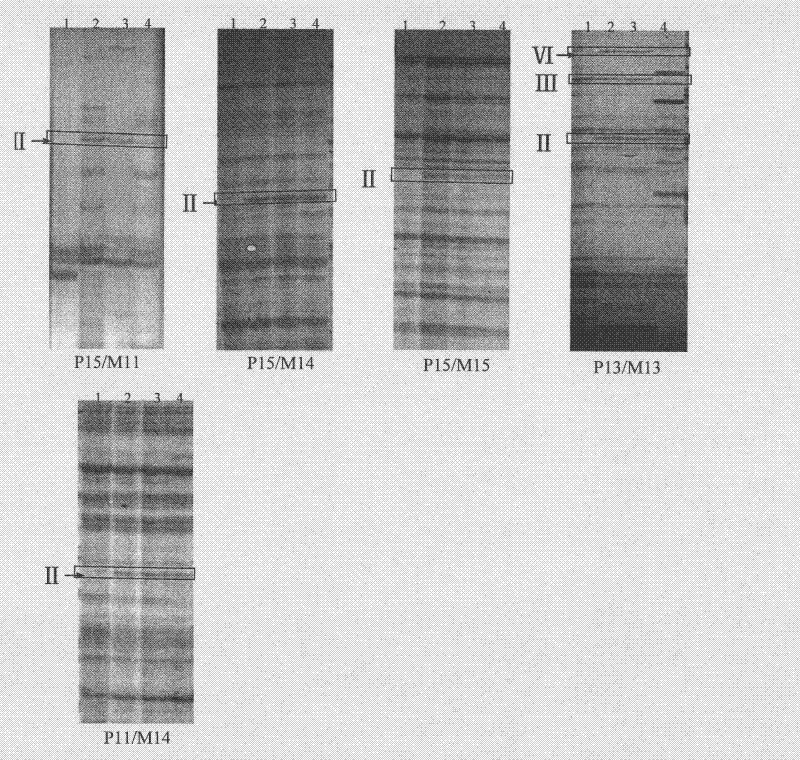
[0040] Embodiment 3, genetic background detection:
[0041] AFLP analysis refers to Vos (Vos, P., R.Hogers, M.Bleeker, M.Reijans, T.van de Lee, M.Hornes, A.Frijters, J.Pot, J.Peleman, M.Kuiper, M. Zabeau. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res, 1995, 21: 4407-4414) and other methods. The adapters and primers used are listed in Table 1.
[0042] 1) Genomic DNA double digestion and adapter ligation: 100×BSA 0.2μl, NEB buffer 4.0μl, Pst I (20U / μl) 0.25μl, Mse I (10U / μl) 0.5μl, Pst I linker (5pM) 1.0μl , Mse I adapter (50pM) 1.0μl, T4 DNAligase 0.4μl, 10×T4buffer 2.0μl, DNA (100ng / μl) 5.0μl, ddH 2 O make up 40 μl. Incubate at 37°C for 8 hours.
[0043] 2) Preamplification: The reaction system is 25 μL, containing 1 μL of ligation product, 0.6 pmol / L primer, 2.5 μL of 10×PCR buffer, 0.2 mmol / L dNTPs, and 1 U Taq polymerase. The PCR amplification program was 94°C for 2min; 94°C for 35S, 56°C for 35S, 72°C for 60S, 30 cycles; 72°C for 5min. The pre-ampl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com