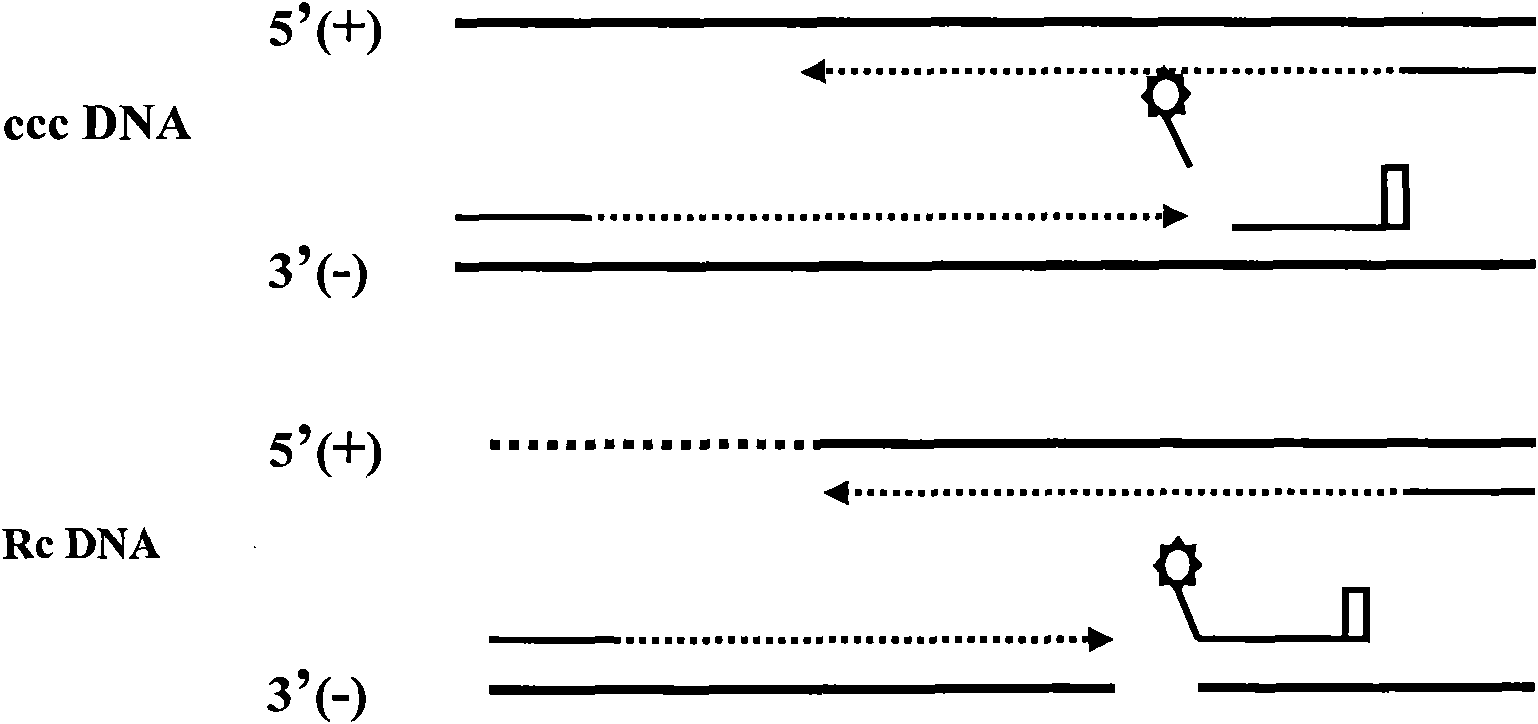
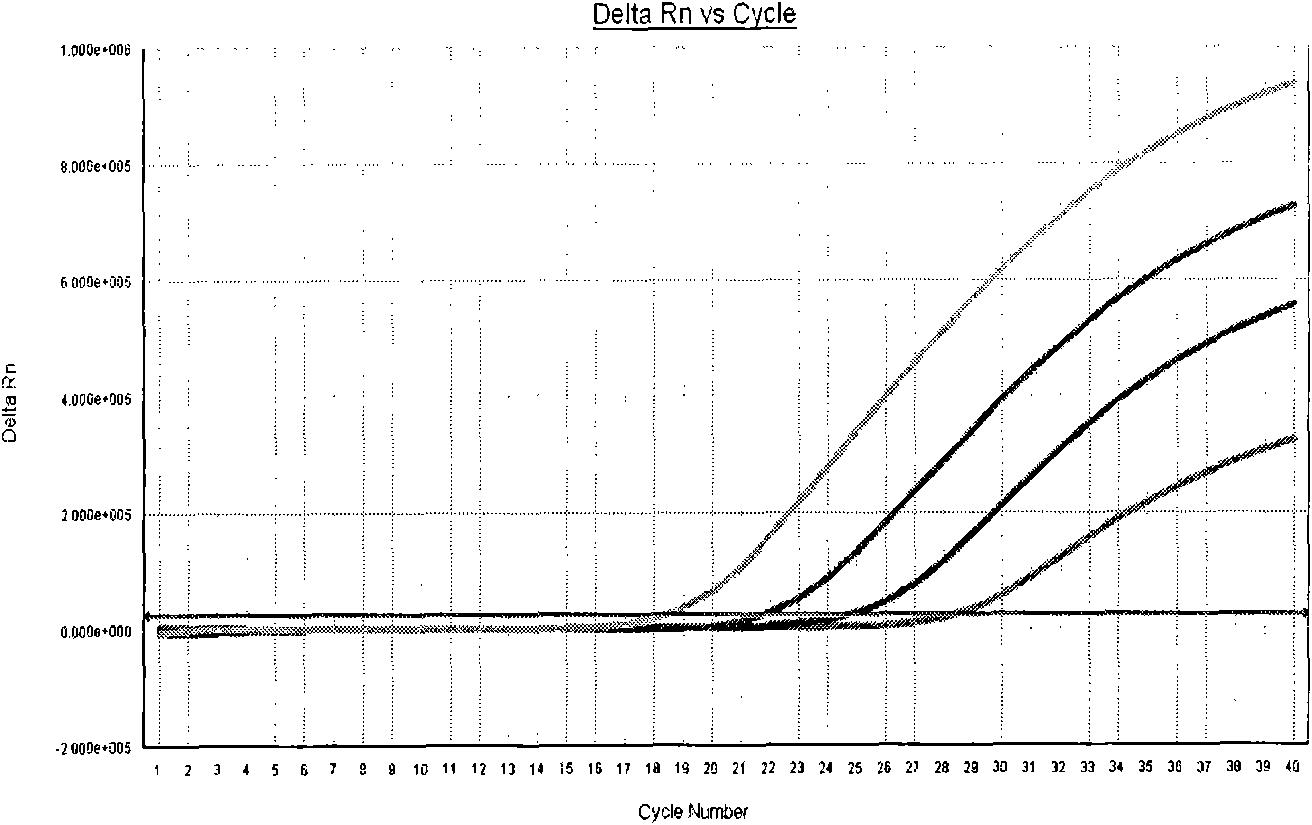
Method for performing two-color fluorescence polymerase chain reaction (PCR) detection on hepatitis B virus covalently closed circular (ccc) DNA, and application of kit
A hepatitis B virus, two-color fluorescence technology, applied in fluorescence/phosphorescence, microbial-based methods, biochemical equipment and methods, etc., can solve problems such as difficult quantification and interference detection results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1: Specimen collection and delivery
[0049] The specimen types of this kit include whole blood and liver tissue, etc. Whole blood can be collected with a sterile syringe to extract 2-3 ml of venous blood from the subject, inject it into a sterile 5 ml sodium citrate anticoagulant tube, invert it repeatedly 3-5 times to fully mix, and store it for testing. For the liver tissue, about 100-500mg of the subject’s liver tissue needs to be taken under aseptic conditions, transferred to a clean 1.5ml centrifuge tube, and stored for testing. Specimens after the above treatment can be stored at -20°C, but not more than 6 months, and can be stored at -70°C for a long time. When sending for inspection, it needs to be stored in a 0°C ice bottle.
Embodiment 2
[0050] Example 2: DNA Extraction
[0051]In order to carry out the method of the present invention, DNA extraction is required. For peripheral blood, first shake the anticoagulated blood, take 500μl and transfer it to a clean 1.5ml centrifuge tube, numbered and marked. Add 1ml 1× red blood cell lysate, shake vigorously for 30s, centrifuge at 3000g for 5min, and discard the supernatant. Repeat twice until there is no obvious red precipitate. If there is still a red precipitate, it is necessary to wash the cell pellet again. Then add 1ml of normal saline, shake vigorously for 15s, centrifuge at 10000g for 5min, and discard the supernatant. Add 50 μl of nucleic acid extraction solution to the precipitate, bathe in 100°C water for 10 minutes, and centrifuge at 13,000 g for 3 minutes. The supernatant is the extracted DNA solution. For liver tissue, it is necessary to wash the blood on the surface of the liver tissue with normal saline, take about 50 mg of tissue, add 50 μl of n...
Embodiment 3
[0052] Embodiment 3: Two-color fluorescent PCR amplification detects HBV cccDNA
[0053] Take out the two-color fluorescent PCR MIX and Taq enzyme system, melt at room temperature, shake and mix well, and then centrifuge at 10,000rpm for 10s. Each test reaction system was prepared as follows:
[0054] Reagent
[0055] Calculate the amount of each reagent used, add it to a clean 0.2ml centrifuge tube of appropriate volume, mix well, centrifuge at 10,000rpm for 10s, add 26μl of the system to the set PCR reaction tube, and then add the processed sample to each tube Or positive quality control or negative quality control 4μl. Before adding the samples, the positive quality control substances were pre-diluted 10 times, 100 times, and 1000 times, and the original solution of the positive quality control substances was marked as 1×10 7 , 1×10 6 , 1×10 5 , 1×10 4 copies / ml. Centrifuge briefly at 10,000 rpm for 30 seconds after adding the sample. Put each reaction tube...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com