PCR method identified by thelephora ganbajun Zang and specific primer thereof
A dry and specific technology, applied in the direction of DNA / RNA fragments, biochemical equipment and methods, microbial measurement / inspection, etc., can solve the problems of insufficient data and high market prices, and achieve good repeatability and strong specificity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] See Table 1 for the collection of Thelephora ganbajun Zang fruiting bodies, pure cultures of Thelephora ganbajun Zang, related species, and associated bacteria.
[0018] Table 1 Fruiting bodies, pure cultures, relative species and associated bacteria of D.
[0019]
Embodiment 2
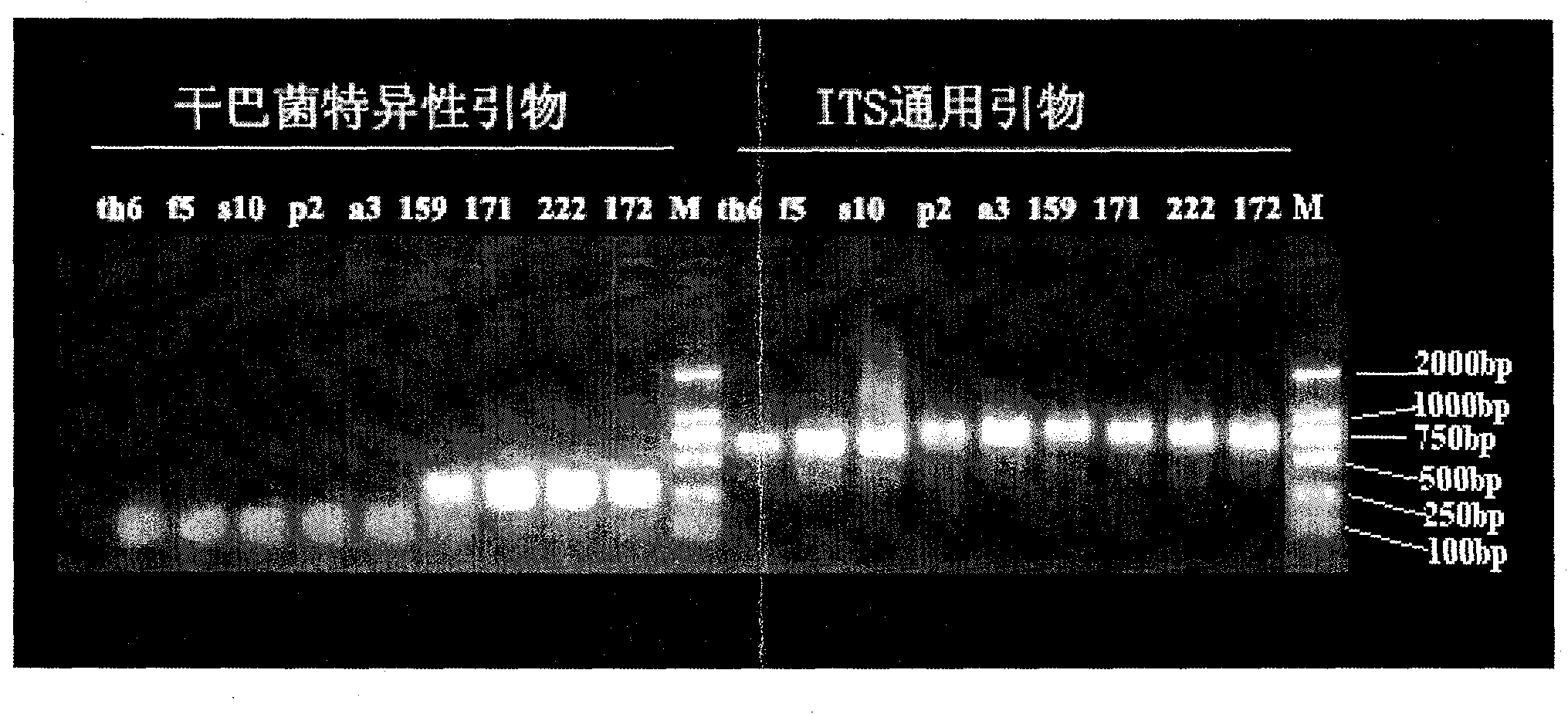
[0021] Specific primer design.
[0022] Search GenBank (http: / / www.ncbi.nlm.nih.gov / genbank / ) for the transcriptional spacer (ITS) sequence of the genus Thelephora, use ClustalX1.8 for sequence alignment, and design according to the variable region The specific primers for Thelephora ganbajun Zang are as follows: Tg1: 5'-ACCTGTGCACCCTCTGTAGTTCC-3'; Tg2: 5'-GTCCAAGCTCATCATGGCA-3'
Embodiment 3
[0024] DNA extraction
[0025] Get about 0.1 g of the fungal samples listed in Table 1 (grown mycelium or fruiting body), transfer to a 1.5 ml centrifuge tube, add 0.30 g of white quartz sand (Sigma) and 60 ° C 2 × CTAB buffer [2 %CTAB (w / v); 100mM Tris-HCl; 1.4M NaCl; 20mM EDTA, pH 8.0] 1ml, grind thoroughly with a plastic mallet; place in a water bath at 60°C for 1 hour, then repeat with phenol:chloroform (1:1) Extract and centrifuge until a clear liquid extract is obtained; use 4°C 100% ethanol to precipitate DNA, wash the DNA pellet with 70% cold ethanol (4°C), dry the DNA pellet with a vacuum centrifugation system, and resuspend the DNA pellet in 100 μl TE buffer solution; take 5 μl DNA solution and electrophoresis on 1.5% agarose gel to detect the purity and integrity of genomic DNA; finally, store the obtained DNA sample at 4°C.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com