Method for identifying disease resistant variety of cruciferous crop
A technology of cruciferous crops and cruciferous, which is applied to the determination/inspection of microorganisms, biochemical equipment and methods, etc., and can solve the problems of long time spent and inadaptability to modern production and construction.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Cloning of XC_0542 Gene and Construction of Expression Vector pBI0542
[0054] According to the gene sequence of XC_0542, design primers:
[0055] Forward primer: F, 5′-GGGTCTAGAATGGAGATCAAGAAAGCGCAGTCC-3′,
[0056] Reverse primer: R, 5'-GGGGGATCCTCAGCCGGAGCGCGGCGGGTC-3'.
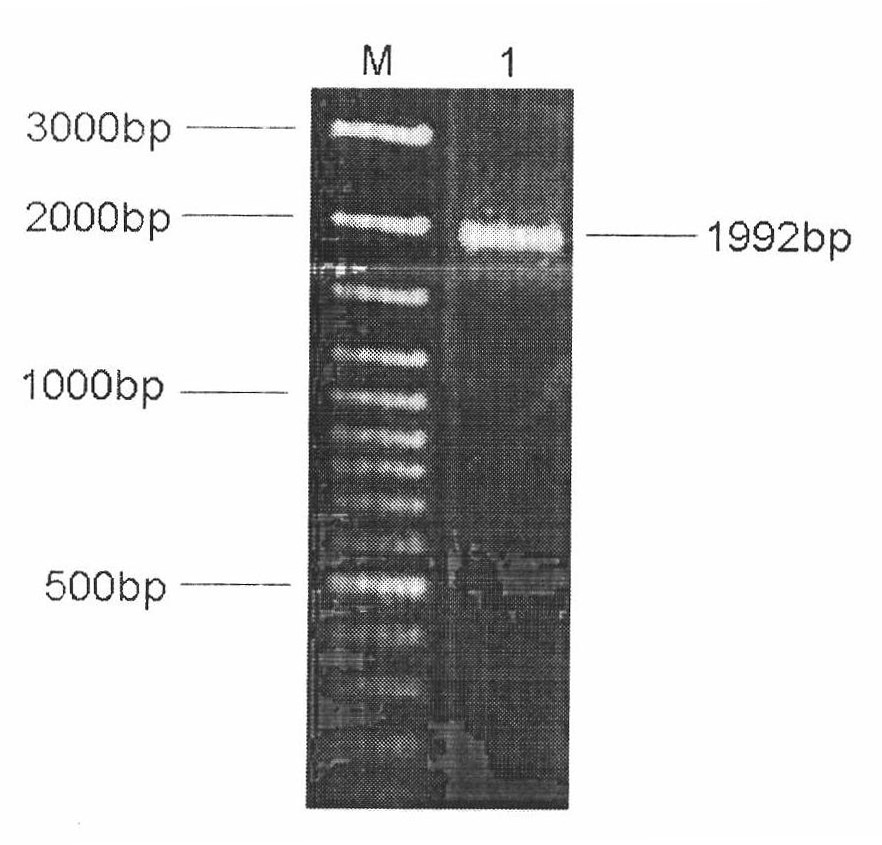
[0057] Using the total DNA of Brassicaceae black rot fungus as a template, the full-length sequence of the gene was amplified by PCR, cloned into pBI121, and the expression vector pBI0542 was constructed and verified by restriction enzyme digestion. Please refer to figure 1 . The sequence was subcloned into pK18mob and verified by sequencing. The sequence was consistent with the sequence of XC_0542 gene (GeneID: 3381550).
[0058] figure 1 Electrophoretic pattern for PCR clone XC_0542 gene, M: 0321 standard DNA (fragment size from large to small: 3kb, 2kb, 1.5kb, 1.2kb, 1.0kb, 0.9kb, 0.8kb, 0.7kb, 0.6kb, 0.5 kb, 0.4kb, 0.3kb, 0.2kb, 0.1kb, ); Lane 1: PCR cloned fragment of XC_0542 gene.
Embodiment 2
[0059] Embodiment 2, introducing expression vector pBI0542 into Agrobacterium
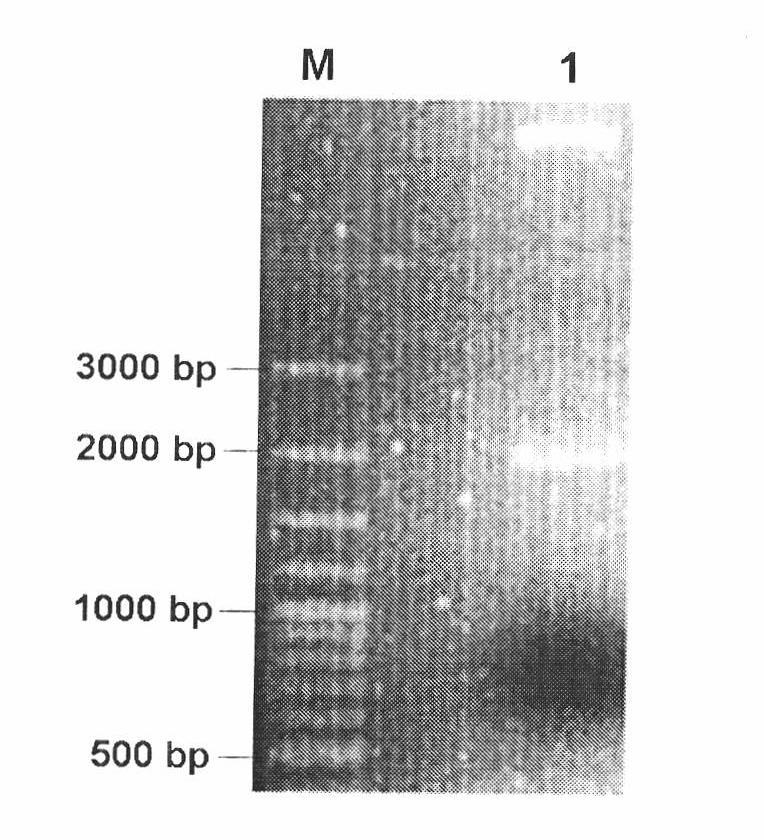
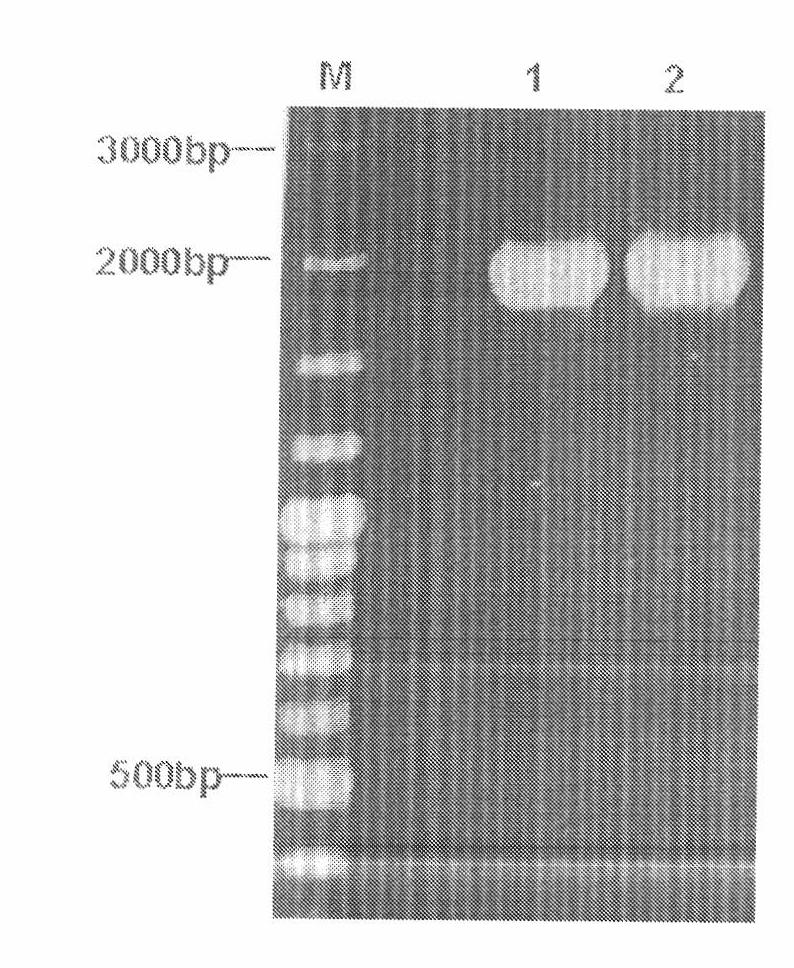
[0060] The triparental pBI0542 was introduced into Agrobacterium tumefaciens EHA105, and the obtained triparental zygote was named EHA105 / pBI0542. For the PCR verification of the triparental zygote, please refer to figure 2 and image 3 .
[0061] figure 2 Restriction electrophoresis pattern of pBI0542 expression vector, M: 0321 standard DNA (fragment sizes from large to small: 3kb, 2kb, 1.5kb, 1.2kb, 1.0kb, 0.9kb, 0.8kb, 0.7kb, 0.6kb, 0.5kb, 0.4kb, 0.3kb, 0.2kb, 0.1kb, ); Lane 1: XbaI / BamHI fragment of gene clone pBI0542.
[0062] image 3 It is the PCR verification electrophoretic pattern of pBI0542 in EHA105, M: 0321 standard DNA (fragment sizes from large to small are: 3kb, 2kb, 1.5kb, 1.2kb, 1.0kb, 0.9kb, 0.8kb, 0.7kb, 0.6kb, 0.5kb, 0.4kb, 0.3kb, 0.2kb, 0.1kb, ); Lane 1: PCR verified fragment of pBI0542 in EHA105.
Embodiment 3
[0064] Agrobacterium Hypersensitivity Reaction (HR) Test
[0065] The plant materials used are cauliflower, A is a disease-resistant variety, and B is a disease-susceptible variety. Healthy cauliflower leaves were selected for testing.
[0066] (1) Inoculate Agrobacterium into LB medium supplemented with corresponding antibiotics and culture overnight at 28°C.
[0067] (2)OD 600 When the temperature is 0.4 to 0.5, collect 1 mL of the bacterial solution in a 1.5 mL EP tube, centrifuge at 5000 rpm for 5 min, pour off the supernatant, and use 2 mL of induction medium (10 mM MgCl 2 , 5mM MES pH5.6, 150μM AS) resuspended, induced at 28°C for 5h.
[0068] (3) Adjust OD with induction medium 600 to 0.4 to 0.5, the wild-type Xcc 8004 of the cruciferous black rot fungus was used as a positive control (such as Figure 4 Middle A1 and B1), EHA105 / pBI121 as negative control (such as Figure 4 in A3 and B3). Use a needle-free syringe to draw the bacterial solution and infuse it into...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com