Luciferase active fragment and application thereof
An active fragment, luciferase technology, applied in the field of biochemistry and cell biology research, can solve the problems of poor stability of enzyme protein, limited application range, less research and reports, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0082] Example 1. Primary sequence analysis of Gaussia luciferase (GLuc) and mass spectrometric identification of disulfide bonds
[0083] The known primary sequence of GLuc was analyzed, and the position of the disulfide bond was confirmed by mass spectrometry.
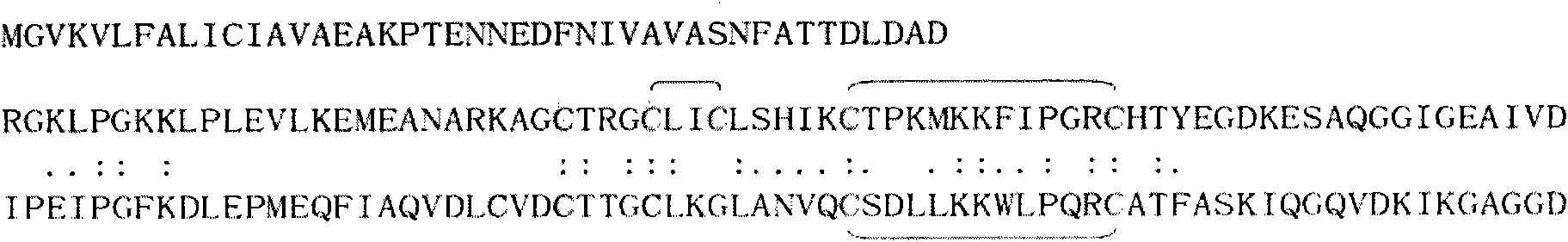
[0084] The test results showed that: the primary sequence of GLuc ( Figure 1A ) contains a total of 185 amino acid residues; its N-terminus is a 17Aa signal peptide sequence, which directs secreted expression; the core enzyme (which ranges from 44 to 185Aa in the sequence) has two highly homologous substructures Each domain contains five cysteine residues; as determined by the Ellman method, GLuc does not contain free cysteine sulfhydryl groups and forms five pairs of disulfide bonds. Further identification by mass spectrometry confirmed three pairs of disulfide bonds: C2-C3, C4-C5 and C9-C10, and the other two pairs of disulfide bonds have not yet been identified.
Embodiment 2
[0085] The artificial synthesis of embodiment 2.Gluc whole gene and Gluc 148DNA sequence, its cloning, expression and target ratio of purification
[0086] According to the codon preference of prokaryotes and eukaryotic mammals, optimize part of the codons of Gluc whole gene and Gluc 148 (Gluc 38-185) DNA sequence, so that it can be expressed efficiently in prokaryotic cells and mammalian cells . The codon-optimized DNA sequences of Gluc 185 and Gluc 148 are shown in Figure 1B In , the comparison of the DNA sequence of Gluc 148 with the original gene sequence is shown in Figure 1C middle.
[0087] The obtained optimized GLuc and GLuc148 genes were respectively cloned into pET-22b(+) plasmid (purchased from Novagen), and the plasmid was used to transform Escherichia coli Origami B strain (purchased from Novagen), and cultivated overnight at 37°C. The culture was inoculated into medium (LB medium), cultured at 37°C for 3 hours, added with IPTG (final concentration 50 μM...
Embodiment 3
[0089] Example 3. Fluorescence emission spectrum of active fragment GLuc 148
[0090] The fluorescence luminescence spectrum of Gluc 148 was studied using a fluorophotometer (available from Varian, model Cary Eclipse). The solution used for testing was 50mM PBS, pH7.8, 500mM NaCl, the concentration of GLuc148, in the reaction system, at room temperature Under the catalysis of coelenterazine (final concentration 1 μM) oxidative luminescence (reaction time 10 seconds). Fluorescence emission spectra were acquired between 400-580 nm.
[0091] Wild-type Gaussia luciferase is based on coelenterazine (Coelenterazine) (see structure Figure 3A ) as the substrate luciferase, catalyzing the oxidation reaction of the substrate (such as Figure 3B shown), the reaction produces CO 2 , and at the same time emit blue light with a wavelength of 480nm. Figure 3C For the luminescence spectrum of the purified active fragment GLuc 148 when it catalyzes the oxidation reaction of coelenteraz...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com