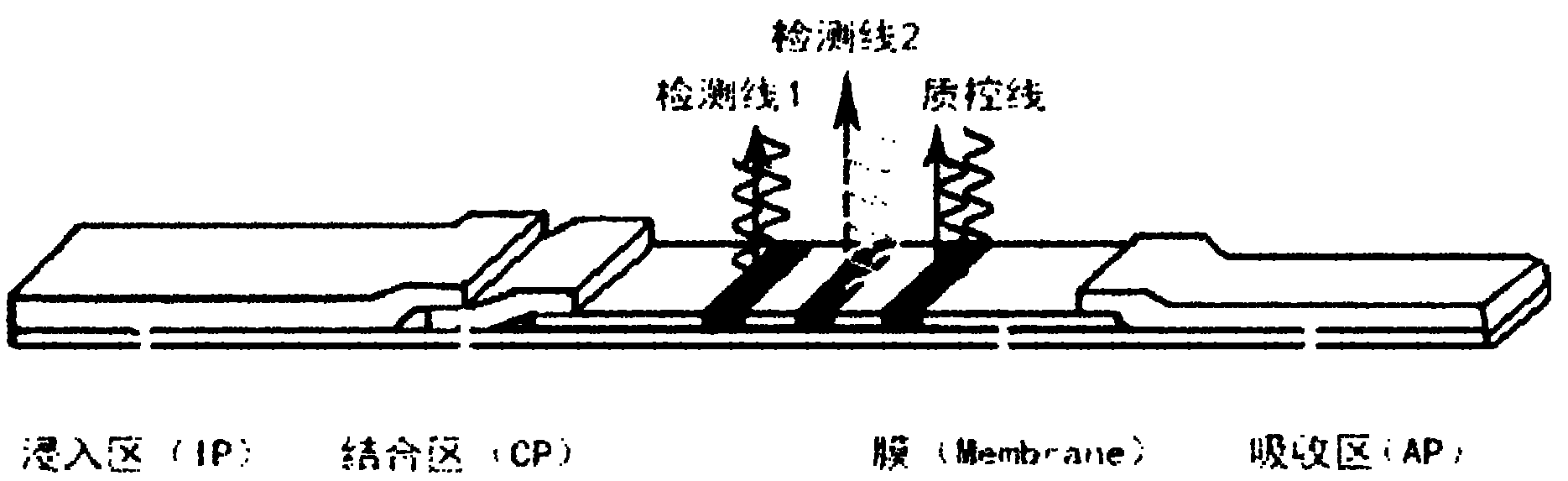
Nucleic acid cross flow test strip-based method for detecting single nucleotide polymorphism
A single nucleotide polymorphism, horizontal technology, applied in the direction of biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of difficult storage, increased cost and steps, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Establishment of a method for detecting KIF single nucleotide polymorphisms based on nucleic acid lateral flow test strips
[0049] (1) Preparation of sulfhydryl-labeled oligonucleotide gold nanoprobes
[0050] ① Preparation of nano-gold colloid
[0051] Prepare a 1mM / L HAuCl4 solution and a 38.8mM / L trisodium citrate solution. Heat the HAuCl4 solution to 130°C and stir it fully during the heating process. After about 20 minutes, quickly add 25mL of the newly prepared trisodium citrate solution (pay attention to heat preservation during this process, do not let the HAuCl4 solution cool down), and continue stirring until the solution After cooling to room temperature, a wine-red solution was formed. Filter the solution with a 0.22 μm nitrocellulose nylon membrane to obtain a nano-gold solution with uniform particles.
[0052] ② Preparation of DNA-labeled gold nanoprobes
[0053] Take 1.0mL of 13nm nano-gold solution with a concentration of 12nM / L, centrifuge at 9500r...
Embodiment 2
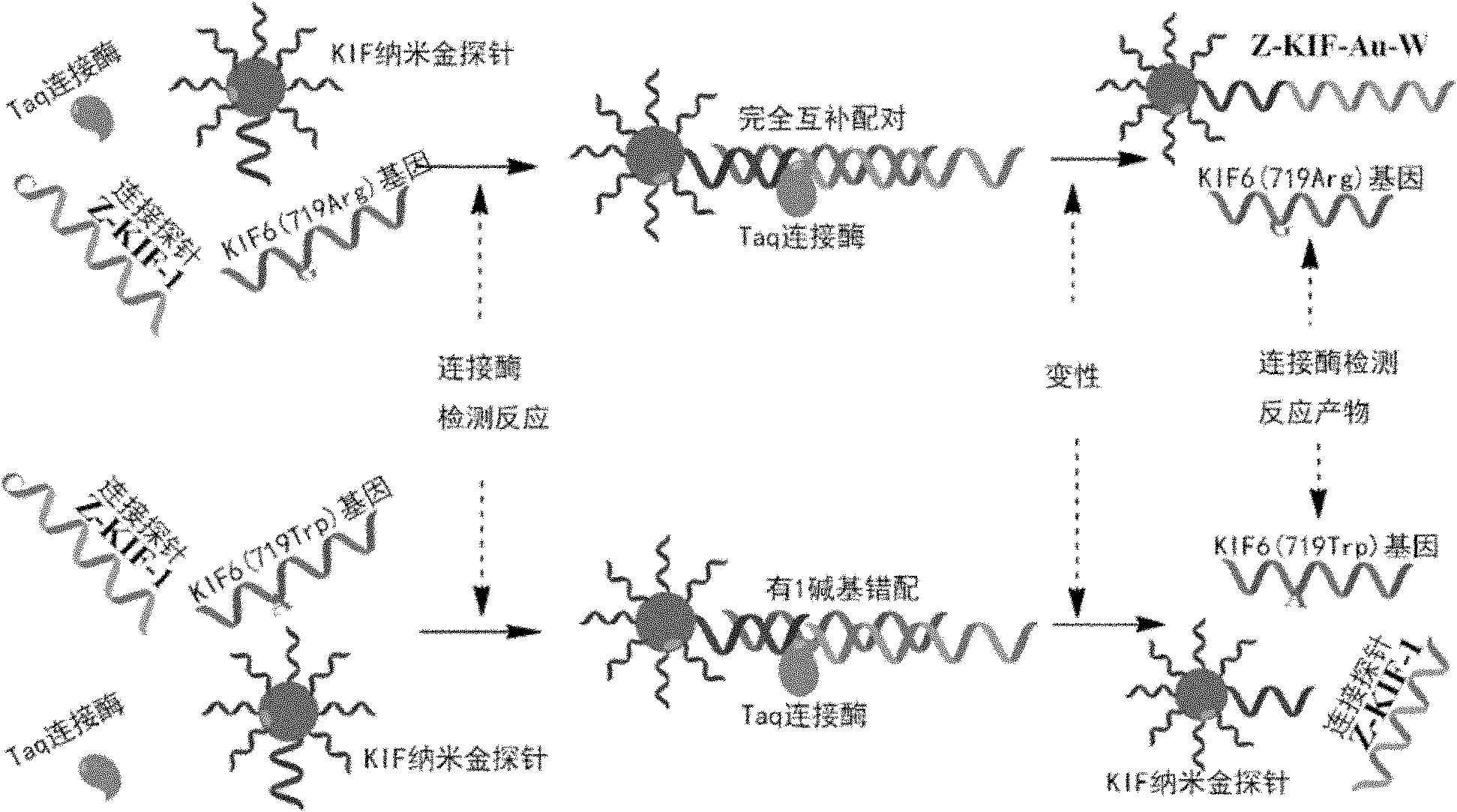
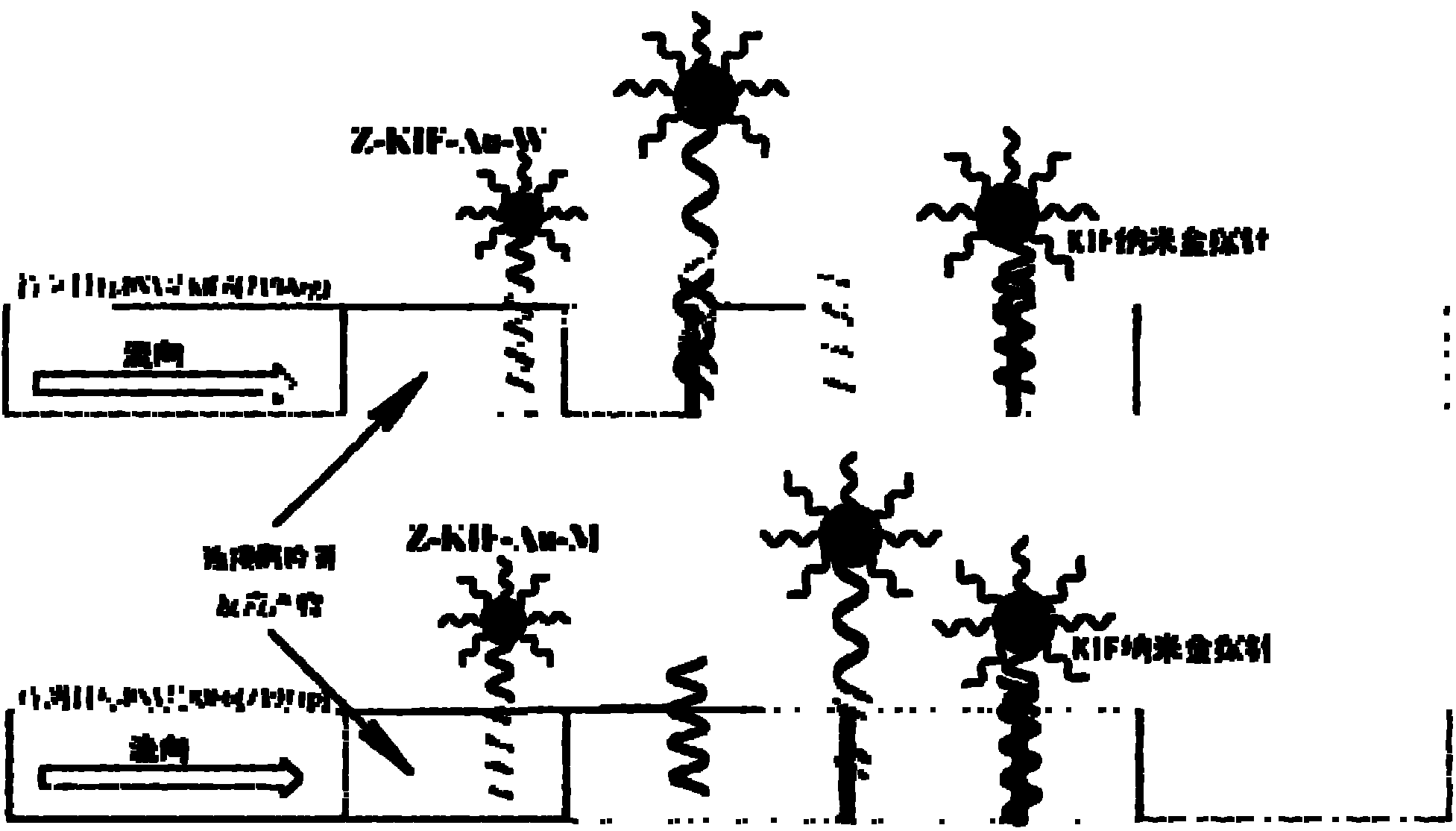
[0074] Detection and judgment of single nucleotide polymorphism in clinical samples of KIF6DNA
[0075] (1) Preparation of sulfhydryl-labeled oligonucleotide gold nanoprobes
[0076] Method is as described in embodiment 1 / (1);
[0077] (2) Design and synthesis of KIF6 DNA detection probe
[0078] The invention adopts a double-probe detection method to design a detection probe and a connection probe according to the DNA sequence of the 719Arg allele (Arg / Trp or Arg / Arg) of the KIF6 gene. Firstly, a sequence (30 bp) containing the single nucleotide polymorphism site of the 719Arg allele of the KIF6 gene was selected as Target DNA, including alleles KIF6 (719Arg) and KIF6 (719Trp). Using the upstream sequence of the single nucleotide polymorphism site as a template, its complementary nano-gold detection probe (KIF-P-PS) was designed, and its 5' end was modified with thiol functionalization to bind to gold nanoparticles. Design two connection probes (27bp), the first base at th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com