Construction method and use of Streptomyces roseosporus gene engineering bacteria
A technology of Streptomyces roseospora and genetically engineered bacteria, which is applied in the field of genetic engineering and microbial fermentation, and can solve the problems of high production cost and low yield of daptomycin
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
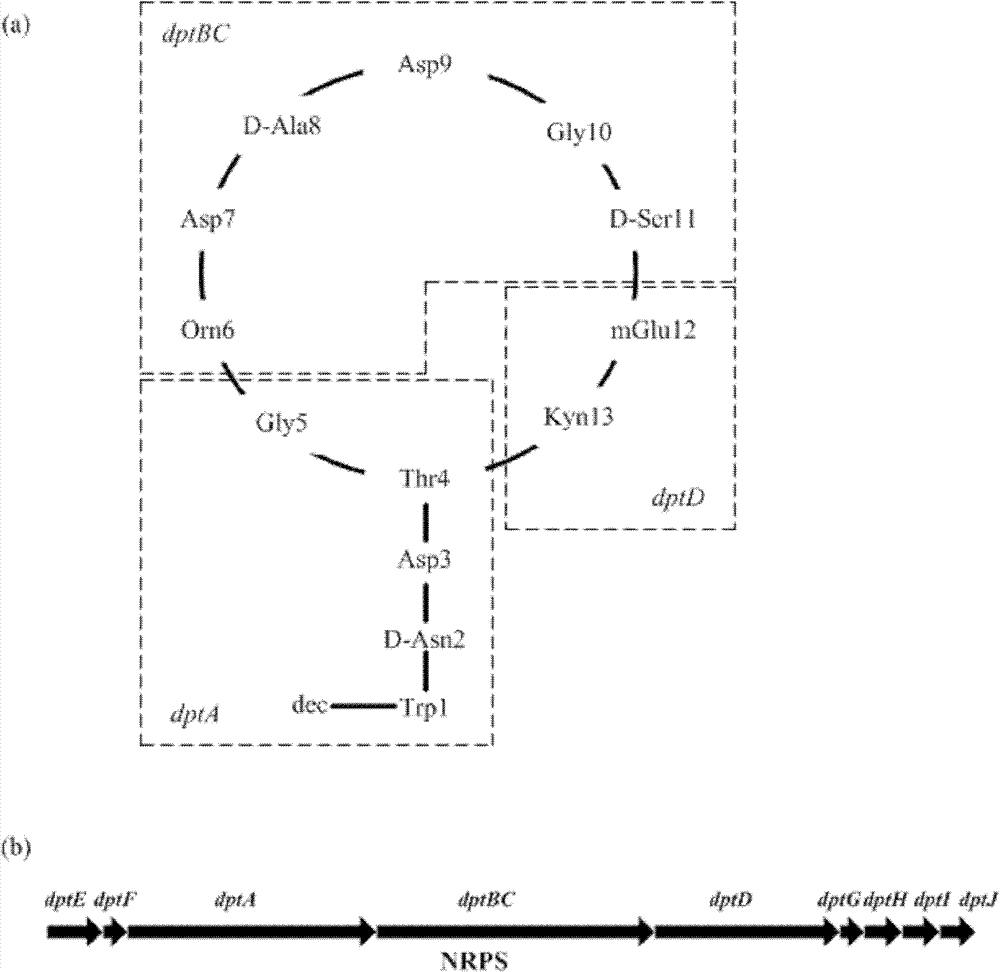
Image
Examples
Embodiment 1
[0027] Construction of Genetic Engineering Bacteria HP-EF01 (CGMCC 3734)
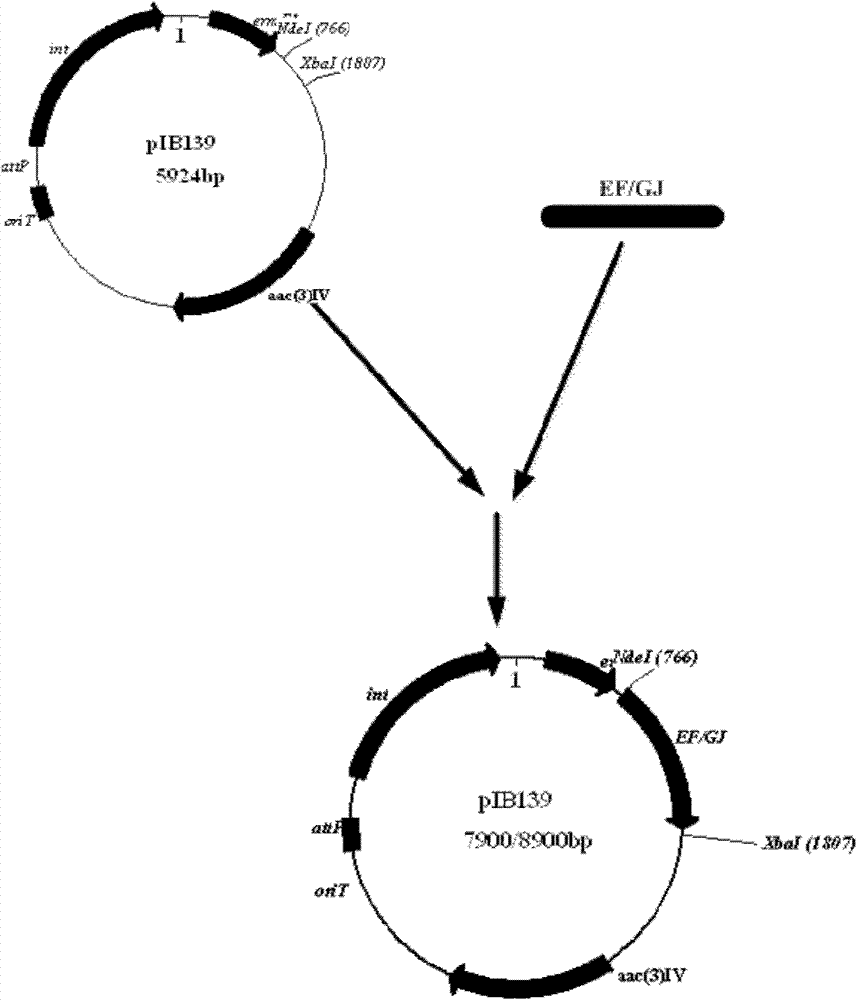
[0028] Primers were designed to PCR clone dptE and dptF (dptEF) located upstream of NRPS. (Upstream and downstream primers are respectively: Upper primer: GGAGTG CATATG GTGAGTGAG (the underline is the NdeI restriction site); Citation: CATCTA TCTAGA ATCCCCTCAGG (the underline is the XbaI restriction site). The PCR product and the expression vector pIB139 were digested with NdeI / XbaI respectively. After recovery, dptEF and pIB139 were ligated with T4 ligase at a ratio of 3:1 to 1:1 at 16°C for 2 hours to construct the recombinant expression vector pEF139 (such as figure 2 ), and then introduce ET12567 Escherichia coli competent cells by the heat shock method, and spread them on the ampicillin-resistant plate after activating and culturing in LB medium for one hour. Then pick the transformant to extract the plasmid and strictly verify it. Then, the recombinant expression vector pEF139 was introduced...
Embodiment 2
[0043] Construction of Genetic Engineering Bacteria HP-GJ01 (CGMCC 3733)
[0044] Design primers to PCR clone dptG, dptH, dptI and dptJ (dptGHIJ) located downstream of NRPS. The upstream and downstream primers are respectively: Top primer: TCCGTG CATATG AGGAAACAT (the underline is the NdeI restriction site); Citation: CGTCAG TCTAGA GACGCTTGC (XbaI restriction site is underlined). The PCR product and expression vector pIB139 were double-digested with NdeI / XbaI respectively, after gel cutting and recovery, dptGHIJ and pIB139 were ligated with T4 ligase at a ratio of 3:1 to 1:1 at 16°C for 2 hours to construct the recombinant expression vector pGJ139 (Such as figure 2 ), and then introduce ET12567 Escherichia coli competent cells by the heat shock method, and spread them on the ampicillin-resistant plate after activating and culturing in LB medium for one hour. Then pick the transformant to extract the plasmid and strictly verify it. Then, the recombinant expression vecto...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com