Liquid phase chip for detecting PIK3CA (phosphoinositide-3-kinase, catalytic, alpha) gene mutation
A detection solution and chip technology, applied in the field of molecular biology, can solve the problems of pollution, poor timeliness, and false positive rate criticism, and achieve the effect of overcoming low sensitivity, improving detection accuracy, and good signal-to-noise ratio.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1PI
[0021] Embodiment 1 PIK3CA gene mutation detection liquid chip mainly includes:
[0022] 1. ASPE Primers
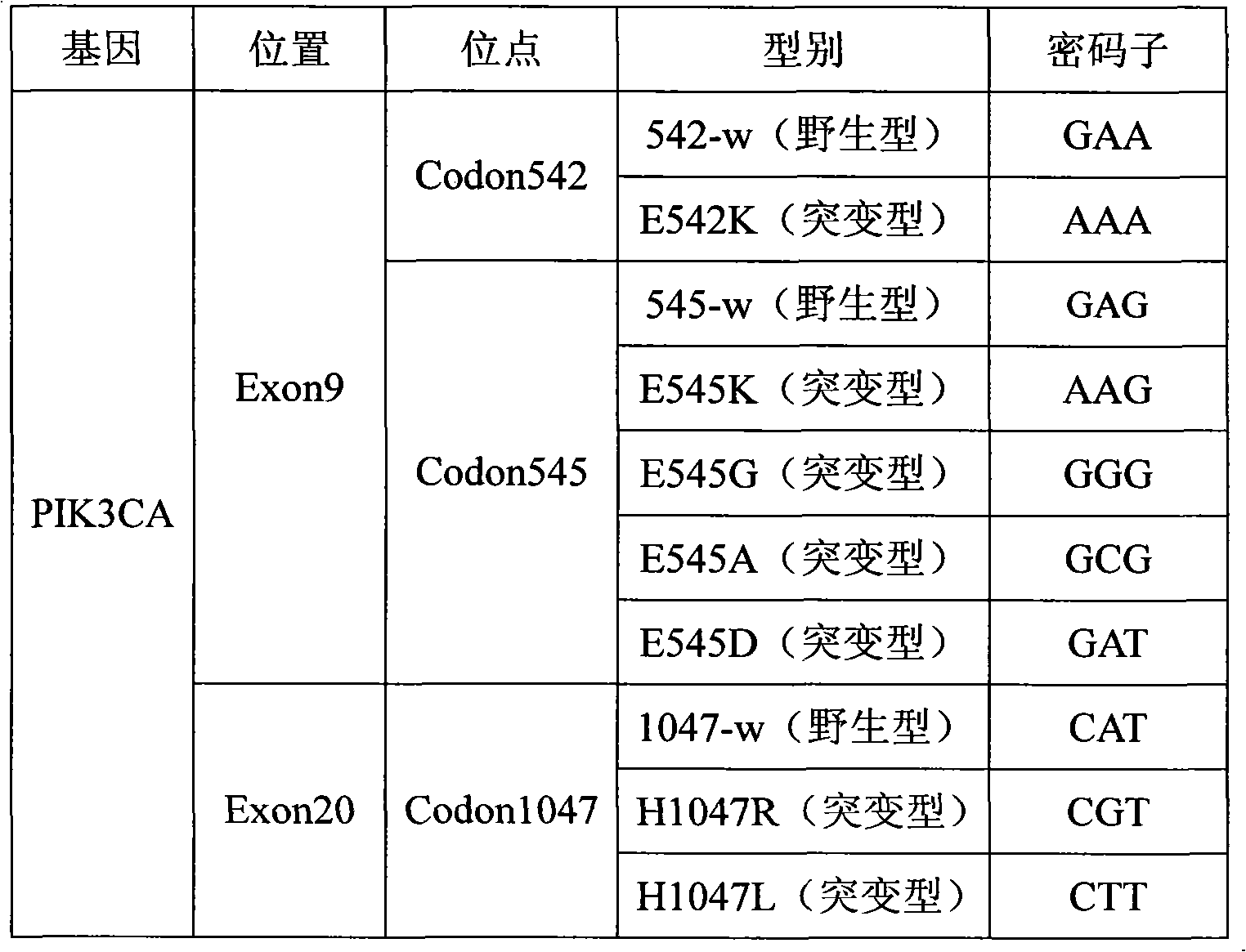
[0023] Specific primer sequences were designed for five mutant types of PIK3CA gene Exon9: E542K, E545K, E545G, E545A, E545D, and two mutant types of Exon20: H1047R, H1047L.
[0024] The design points of ASPE primers for PIK3CA gene mutation detection are as follows:
[0025]ASPE primers consist of "Tag sequence + specific primer sequence". Among them, the 5' end is the Tag sequence designed according to the PIK3CA gene mutation detection. The designed Tag sequence can avoid the secondary structure that may be formed by the ASPE primer in the reaction system to the greatest extent, and the Tag sequence and the Tag sequence, the Tag sequence and the specific There is no cross-reaction between the primer sequences. The Tag sequence and the specific primer sequence form complete ASPE primers, and enable all ASPE primers to react synchronously in a uniform reaction system ...
Embodiment 2
[0052] Example 2 Detection of samples using PIK3CA gene mutation detection liquid chip
[0053] The formula of described various solutions is as follows:
[0054] 50mM MES buffer (pH5.0) formula (250ml):
[0055] Reagent
[0056] 2×Tm hybridization buffer
[0057] Reagent
[0058] Store at 4°C after filtration.
[0059] ExoSAP-IT kit was purchased from US USB Company.
[0060] Biotin-labeled dCTP was purchased from Shanghai Sangon Bioengineering Technology Service Co., Ltd.
[0061] 1. Sample DNA extraction:
[0062] Refer to the relevant methods of DNA extraction in "Molecular Cloning" to obtain the DNA to be detected.
[0063] 2. PCR amplification of samples to be tested
[0064] Using Primer5.0 to design primers, multiplex PCR amplifies the target sequence with detection sites in one step, and the product sizes are 141bp and 108bp, respectively. The primer sequences (SEQ NO.81-84) are shown in Table 7 above.
[0065] First prepare the PCR primer ...
Embodiment 3
[0117] The selection of wild-type and mutant-type specific primer sequences with different source sequences in embodiment 3
[0118] 1. Design of liquid-phase chip preparation (selection of wild-type and mutant-specific primer sequences)
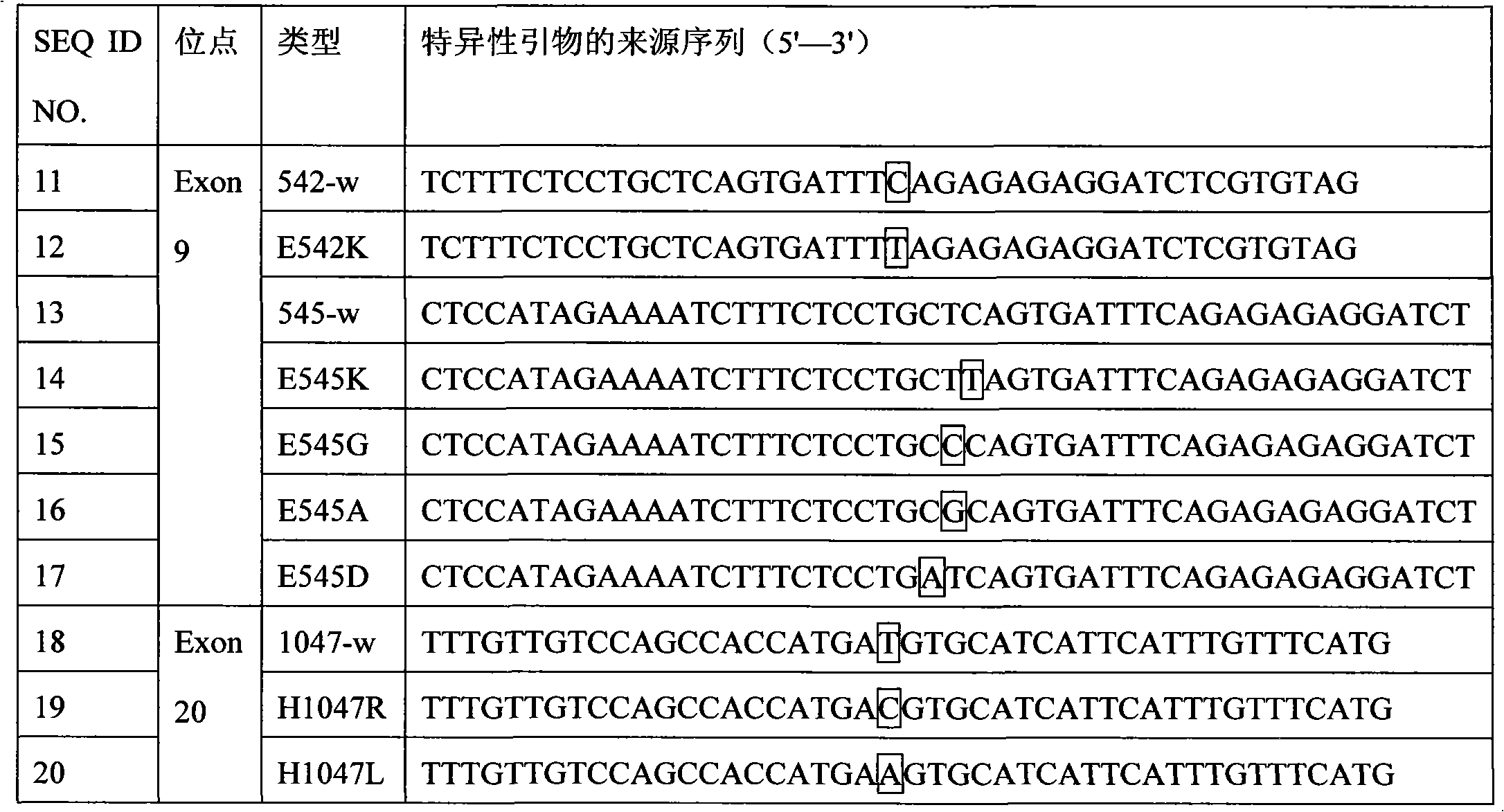
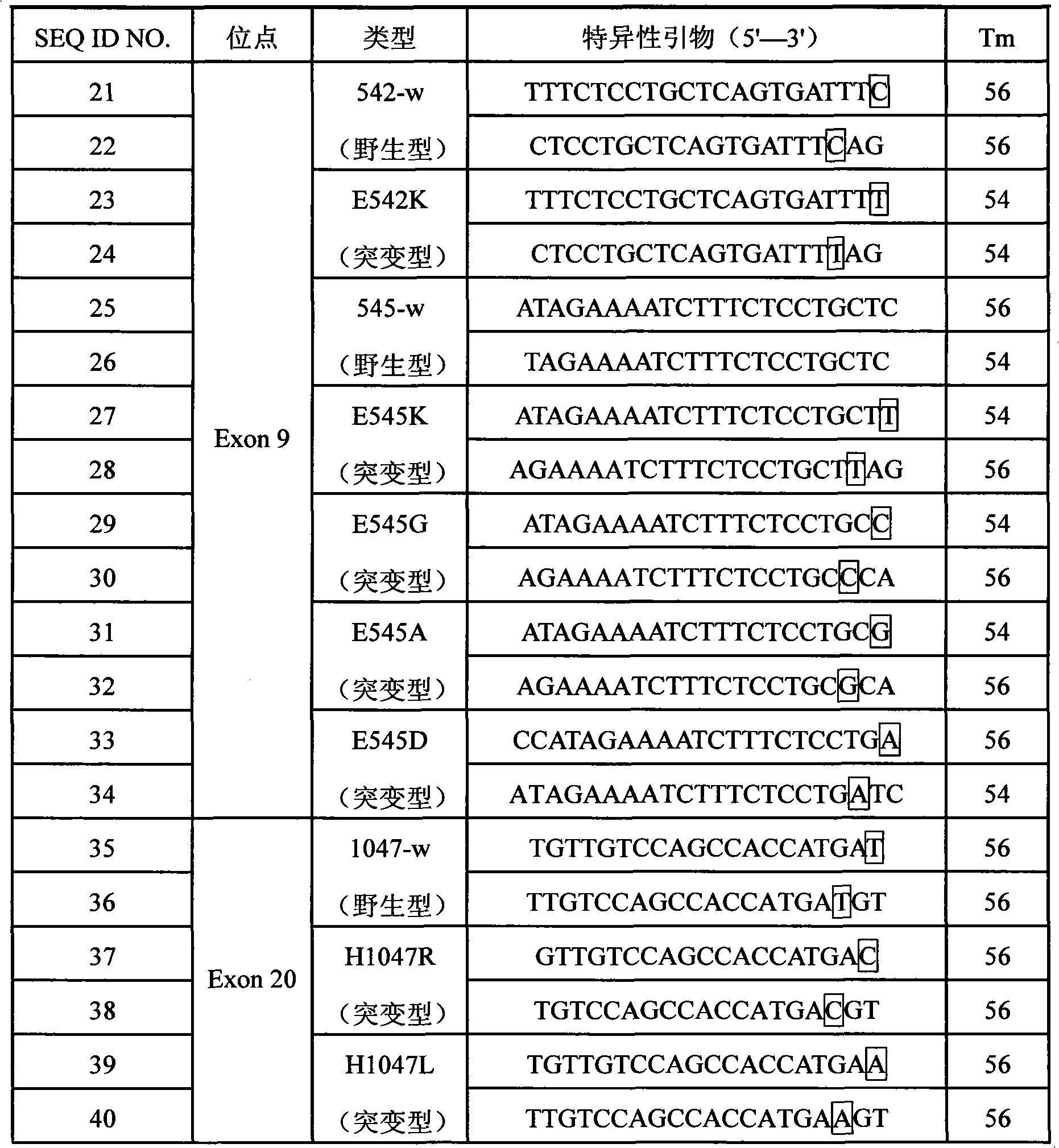
[0119] For the PIK3CA gene Exon9 and Exon20 mutation sites, the specific primer sequences of the 3' end of the wild-type and mutant ASPE primers were designed respectively, and the specific primer sequences for detecting Exon9 and Exon20 mutations were derived from the SEQ ID of SEQ.NO11-20 NO.21-SEQ ID NO.40, or SEQ ID NO.51-SEQ ID NO.70 derived from SEQ NO.41-50. The Tag sequences at the 5' ends of the wild-type and mutant ASPE primers are respectively selected from SEQ ID NO.1-10, and correspondingly, the anti-tag sequences coated on the microspheres that are complementary to the corresponding tag sequences are respectively selected from SEQ ID NO. .71-80. The specific design is shown in the following table (Table 12). The synthesis of...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com