A method for identification of distant hybrid offspring of peony and peony
An identification method and a distant hybridization technology are applied in the identification field of the distant hybrid progeny of peony and peony, and can solve the problems of scarcity, low hybrid germination rate, long hybridization time and the like.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
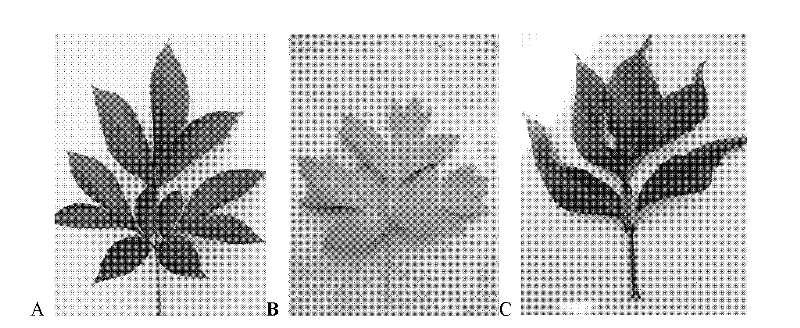
[0028] Embodiment 1 Hybrid Seedling Selection and Morphological Character Investigation
[0029] Method for obtaining hybrid seedlings: pick the flower buds of the male parent back during the dew stage, pick off the anthers indoors, and dry them in the shade for 24 hours in a dry place without direct sunlight. Gently tap the anthers until the pollen is completely scattered, separate the pollen and anthers with a sieve, collect the sieved pollen in a clean vial, label it and store it in a -20°C refrigerator for later use. Select a robust plant as the female parent, use tweezers or bare hands to pinch off or tear off the petals and stamens in the female parent's flower bud stage or dew stage, cover the flower head with a sulfuric acid paper bag and squeeze the bag tightly with a paper clip. After 3-4 days of female parent bagging, pollination begins when the stigma begins to secrete mucus. Pollination was carried out before 11:00 or after 17:00, repeated for three consecutive d...
Embodiment 2
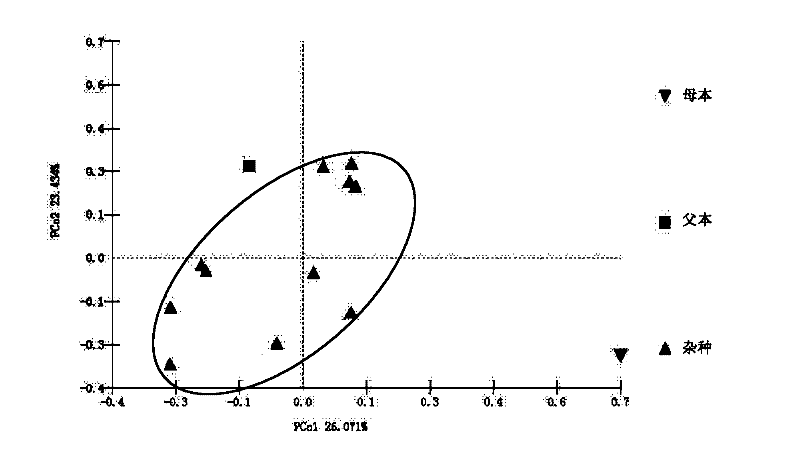
[0039] Embodiment 2 Amplification and sequencing of chloroplast genes
[0040] After the young leaves of 21 hybrid offspring and 2 parents in Example 1 were dried on silica gel, the total DNA was extracted using the Plant Genome Extraction Kit of Tiangen Biotechnology Company, and the operation was performed according to the kit instructions.
[0041] Two chloroplast gene fragments petB-petD and rps16-trnQ were selected for sequencing. The amplification primers of petB-petD are shown in SEQ ID No.1 and 2; the amplification primers of rps16-trnQ are shown in SEQ ID No.3 and 4.
[0042] The polymerase chain reaction (PCR) was completed on the My CyclerTM Thermal Cycler type PCR instrument of BIO-RAD Company. The amplification system is a 25 μl system: template 20-30ng, 10×PCR buffer 2.5 μl, dNTP 80mmol / L, primer 5umol / L, Taq enzyme 1U (Beijing Zexing Biotechnology Co., Ltd.) plus double distilled water to 25ul. The amplification procedure is:
[0043] Pre-denaturation at 94°C...
Embodiment 3
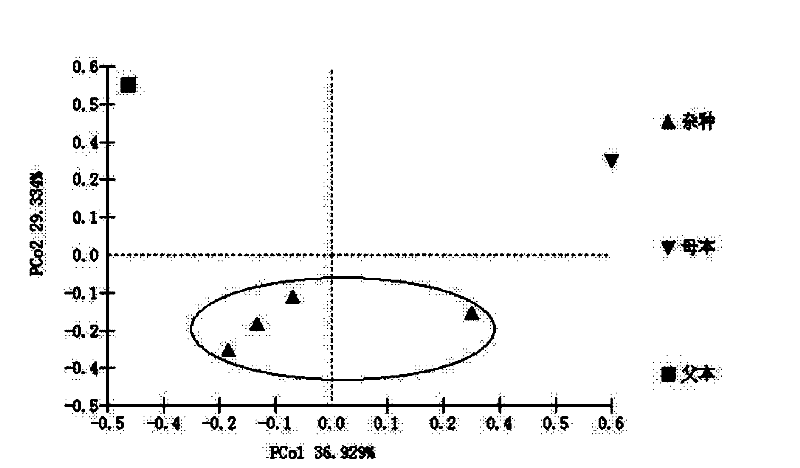
[0056] Amplification and sequencing of embodiment 3SSR markers
[0057] From the 14 pairs of primers screened by Yuan Junhui (his doctoral thesis: The origin of peony and Yan'an peony 2009.12), 10 pairs of primers with high polymorphism and which can be used for experimental samples were selected for amplification (Table 3) (SEQ ID No.5~SEQ ID No.24).
[0058] Table 3 SSR primers
[0059]
[0060] The 5' end of the upstream primer of each pair of primers was fluorescently labeled, wherein Pdel05, Pdel20, Pdel29b, and Pdel33 were labeled with FAM, Pdel02-2, Pdel07, Pdel22, and Pdel35 were labeled with HEX, and Jx02-2 and Jx17 were labeled with ROX.
[0061] The polymerase chain reaction (PCR) was completed on the My CyclerTM Thermal Cycler type PCR instrument of BIO-RAD Company. The amplification system is a 20 μl system: template 20-30ng, PCR Mix (Beijing Biotech Biotechnology Company) 10 μl, 5 μmol / L primers 1 μl each, and double distilled water 20 μl.
[0062] The ampl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com