Specific detection method and kit for trace amount of breast cancer cells in circulating blood
A breast cancer cell and detection method technology, which is applied in the field of specific detection and kits for trace breast cancer cells in circulating blood, can solve the problems of high environmental purity requirements and high false positive rate of antibody specificity requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Example 1 Preparation of MAM mRNA samples and their controls for detection
[0077] MDA-MB361 cells were cultured in vitro, the adherent cells were digested with trypsin to suspend the cells, the culture medium was removed by centrifugation, and then normal saline was added to prepare a single cell suspension, and a certain amount of cells (10 7 )conduct experiment. According to the manufacturer's instructions, total cellular RNA (positive for MAM mRNA expression) was extracted from MDA-MB361 cells with the total cellular RNA extraction reagent, and the optical density (OD value) at 260 and 280 nm was measured to quantify the amount of total RNA. Then the total RNA was serially diluted with TE buffer (pH 7.2), and equal volumes of solutions of each dilution contained 5 (Group A), 10 4 (Group B), 10 3 (Group C), 10 2 (Group D) and 10 1 (Panel E) The amount of total RNA in MDA-MB361 breast cancer cells.
[0078] According to the above m...
Embodiment 2
[0080] Example 2 Synthesis and preparation of aptamers
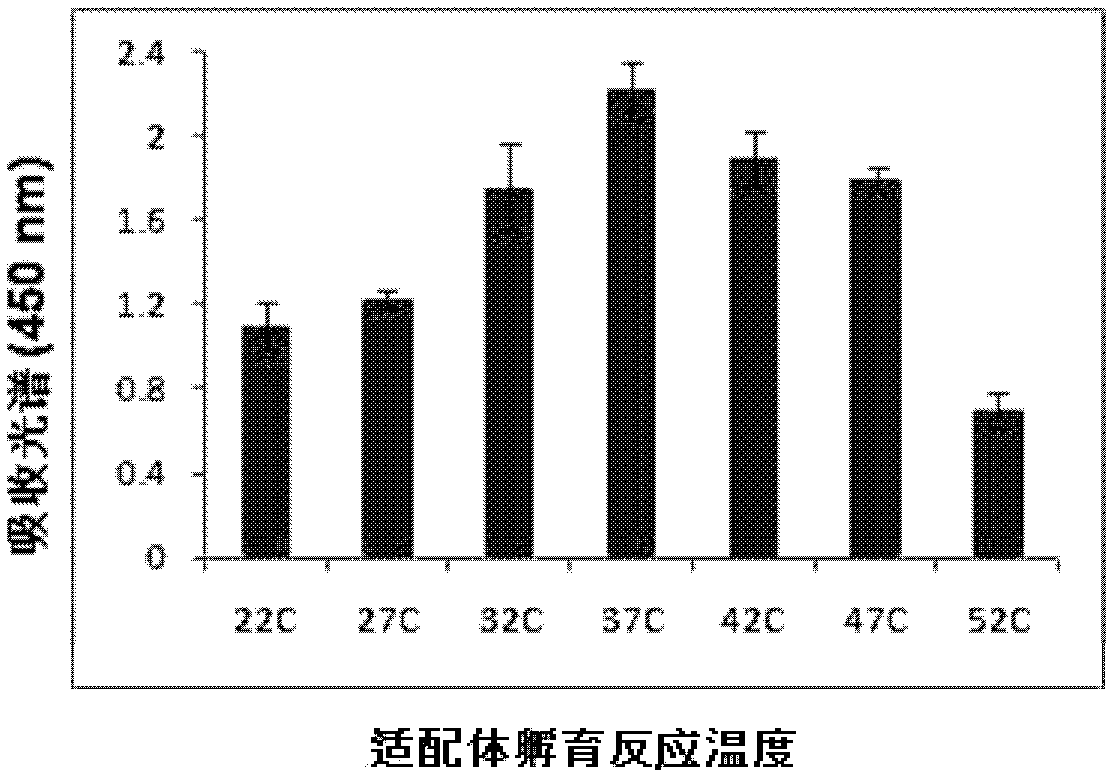
[0081] Take 12 microliters of the carboxy-terminal magnetic body solution and place it in a 1.5ml centrifuge tube, wash it three times with 0.1M imidazole buffer (each time add 150 microliters of 0.1M imidazole buffer (pH7.0) and mix well, then centrifuge. Discard the supernatant), add 300 μl of 0.1M imidazole buffer (pH7.0) containing 0.03M EDC, shake the centrifuge tube slowly for 20 minutes, and then add 12 pmol of the first aptamer (5'-aaggaagccgctgtc-3 ') was added, mixed and incubated at 37°C for 1 hour, during which the centrifuge tube was continuously shaken slowly. Place the centrifuge tube on a magnetic separator to apply magnetic force, blot the supernatant and wash it three times with washing solution (add 300 μl of washing solution (PBS solution containing 100mM NaCl, pH 7.2) and blot dry) for three times. Unbound free first aptamer was removed, and then dissolved by adding 240 μl of dissolving solution...
Embodiment 3
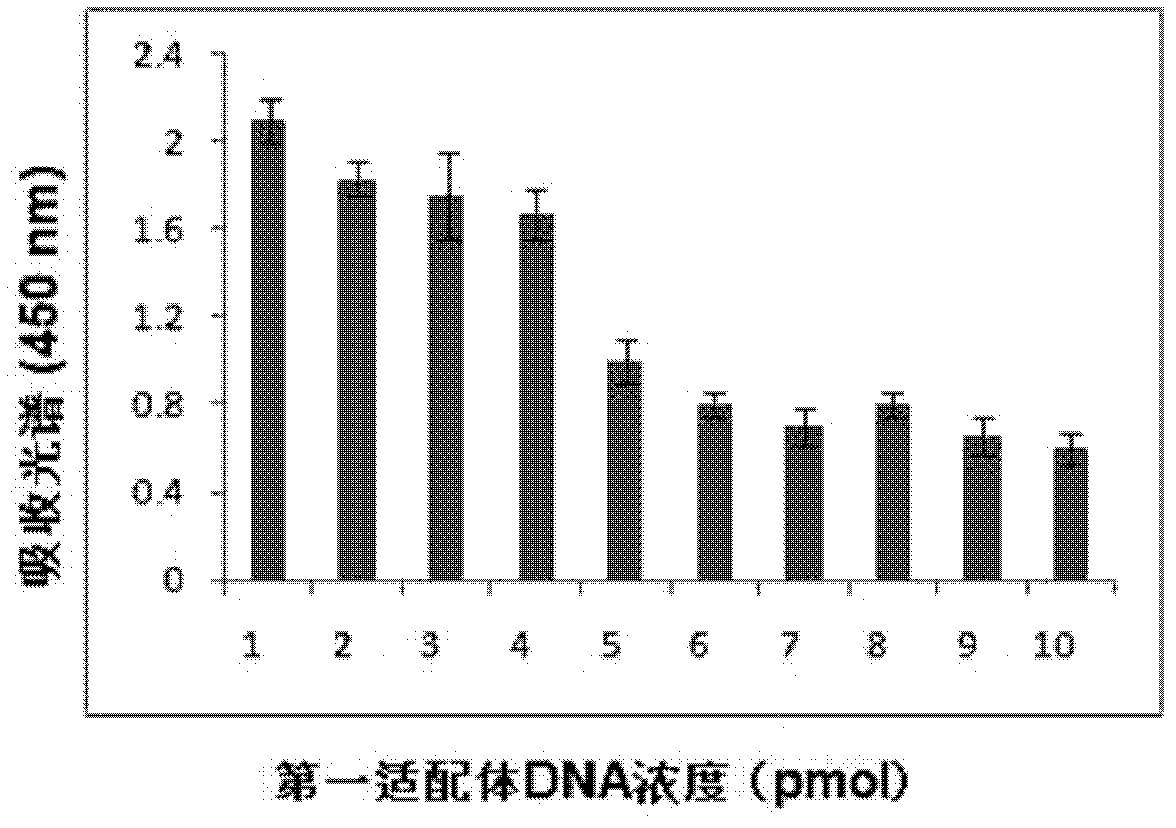
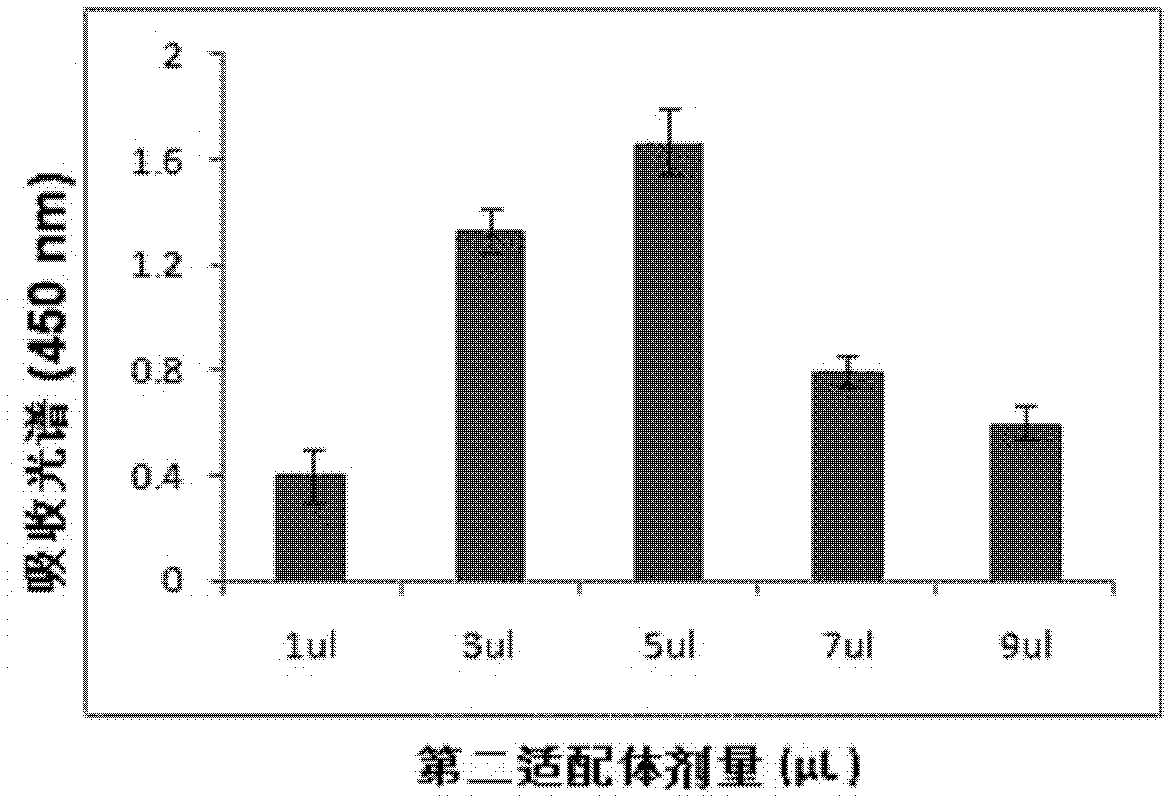
[0084] Example 3 Determination of the concentration of the first aptamer and the second aptamer
[0085] In a series of reaction tubes, total RNA (1 μg) of MDA-MB361 cells known to contain the target molecule (MAM mRNA) was added, together with the first aptamer coupled to magnetic particles prepared according to Example 2 (2- 20 μl, 1-10 pmol), add PBS (0.1M, pH7.2) to 100 μl, incubate at 37°C for 60 minutes; then place the test tube on a magnetic separator to apply magnetic force, and blot the supernatant solution and washed three times with washing solution (300 μl of washing solution (PBS solution containing 100mM NaCl, pH 7.2) was added each time and blotted dry), adding 95 μl of PBS buffer, mixing, and then adding 5 μl according to the implementation The second aptamer coupled with gold nanoparticles prepared in Example 2 was incubated at 37°C for 60 minutes, and then the test tube was placed on a magnetic separator to apply magnetic force, the supernatant was blotted...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com