Direct PCR (Polymerase Chain Reaction) method without extracting animal tissue DNA (Deoxyribonucleic Acid)
A technology of animal tissue and tissue, applied in the field of PCR amplification, to achieve the effect of rapid cost, cost saving and simple method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Configure four different tissue lysates:
[0022] Lysis buffer 1: 10mM Tris-Cl pH8.0, 1mM EDTA pH8.0, 200μg / ml proteinase K;
[0023] Lysis solution 2: 10mM Tris-Cl pH8.0, 1mM EDTA pH8.0, 200μg / ml proteinase K, 0.1% SDS;
[0024] Lysis buffer 3: 10mM Tris-Cl pH8.0, 1mM EDTA pH8.0, 200μg / ml proteinase K, 0.01M NaCl;
[0025] Lysis solution 4: 10mM Tris-Cl pH8.0, 1mM EDTA pH8.0, 200μg / ml proteinase K, 0.1% SDS, 0.01M NaCl;
[0026] Take 0.01g of ear tissue, put it into a clean 200μl PCR tube, shred it fully, then take 50μl of the above four different lysates, first incubate at 55°C for 30min, and then denature at 95°C for 5min to inactivate proteinase K. Finally, 2 μl of four different digestion solutions were directly taken as templates, and the ear tissue without any lysate treatment was used as a control for PCR amplification to amplify the porcine myhc IIb gene fragment.
[0027] PCR reaction system: final concentration of 250μM dNTP, 10mM Tris-Cl, pH8.3, 50mM KCl,...
Embodiment 2
[0031] Lysis buffer: 10mM Tris-Cl pH8.0, 1mM EDTA pH8.0, 100μg / ml proteinase K, 0.01M NaCl
[0032] Take 0.01g of ear tissue, put it into a clean 200μl PCR tube, shred it fully, then take 50μl of the above-mentioned lysate, incubate at 37°C for 25min, then denature at 95°C for 5min to inactivate proteinase K, and finally directly Take 2 μl of digestion solution as a template, and use ear tissue without any lysate treatment as a control for PCR amplification to amplify the porcine myhc IIb gene fragment. The PCR reaction system and reaction conditions are the same as in Example 1.
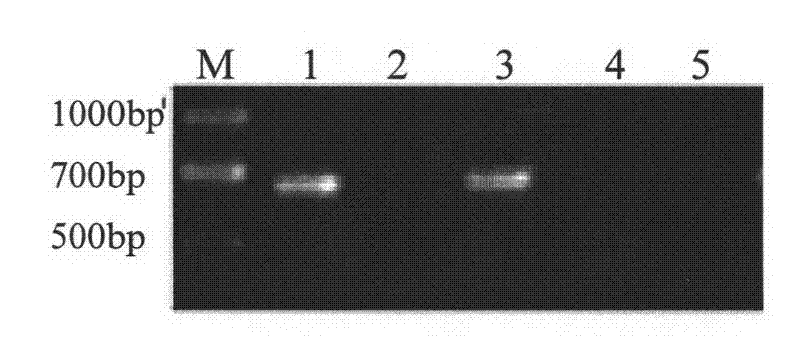
[0033] After the amplification, 5 μl of the amplification product was taken for agarose gel analysis. It was found that a fragment of about 700 bp could be amplified from the ear tissue digested with the lysate, but no band could be amplified from the undigested ear tissue.
Embodiment 3
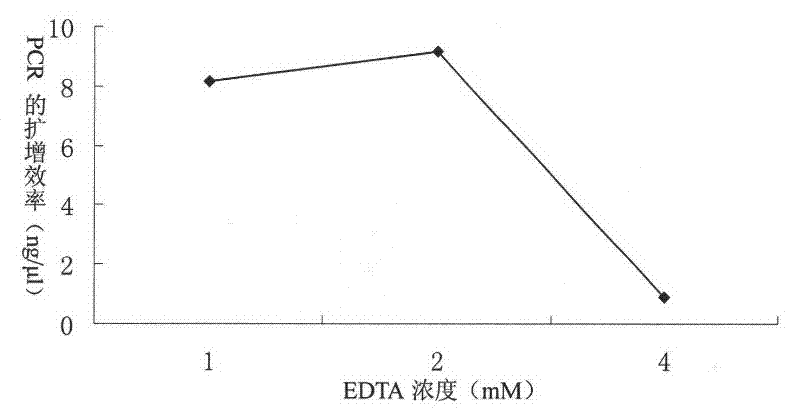
[0035] Optimize the concentration of EDTA in lysate 1: 10mM Tris-Cl, pH8.0, 1mM EDTA, pH8.0, 200μg / ml proteinase K, settings 1, 2, 4, 6, 8, 10, 12, 14, 11 different concentrations of 16, 18, 20mM. The treatment method of the ear tissue and the PCR reaction system and conditions are the same as in Example 1. Three treatments were performed for each concentration, and three repeated PCR amplifications were performed for each treatment to amplify the porcine myhc IIb gene fragment. After the PCR product was detected by agarose gel, it was found that no band could be detected after the concentration of 6mM ( figure 2 ). The PCR products of the first three concentrations were purified by agarose gel electrophoresis, and the purified products were determined by NanoDrop 2000 (NanoDrop Technologies, Wilmington, DE, USA). The results show that 2mM EDTA is the optimal concentration ( image 3 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com