Serum miR-21 (micro-ribonucleic acid-21) detection kit based on luorescence quantitative PCR (polymerase chain reaction) and application thereof
A fluorescence quantitative detection, mir-21 technology, applied in the direction of fluorescence/phosphorescence, microbial measurement/inspection, DNA/RNA fragments, etc., can solve the problems of reducing detection accuracy, cost reduction, increasing non-specificity of reaction, etc., and achieve cost Low cost, easy operation and stable performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Embodiment 1Trizol reagent is prepared
[0060] Trizol reagent preparation, first mix 100g of guanidine isothiol, 118mL of distilled water, 7.04mL of 0.75M sodium citrate, 10.56mL of 10% sodium lauryl sarcosinate, and 20mL of 2M sodium acetate, stir and dissolve; Put 200mL of redistilled phenol (crystal at room temperature) in a water bath at 60°C until completely melted, then add 0.2g of 8-hydroxyquinoline and 1.2g of β-mercaptoethanol or 0.5g of dithiothreitol; then mix all the above reagents , stirred thoroughly, and finally adjusted the pH value to 4.6, and stored at 4°C. The shelf life is about 9 months.
Embodiment 2
[0061] The extraction of embodiment 2 serum RNA
[0062] (1) Take 33 cases of breast cancer patients and 49 cases of healthy people and put them into 2mL EP tubes that have been treated with DEPC water, then add 1mL Trizol reagent prepared in Example 1, and 200 μL chloroform, vortex fully Homogenize for 1 min (note that if the vortex time is not enough or the strength is not strong, the layers will not be well layered). Then let stand for 3min.
[0063] (2) Centrifuge the EP tube at 12000 rpm at 4°C for 15 minutes. Carefully pipette 600 μL of the upper aqueous phase into a 1.5mL EP tube (do not touch the white layer in the middle, otherwise it will easily lead to protein contamination). Then add 600 μL of pre-cooled isopropanol, mix thoroughly, and centrifuge at 4°C for 15 minutes.
[0064] (3) Pour off the centrifuged liquid carefully. The remaining liquid can be centrifuged for a while and then sucked up with a 200mL gun, but try not to touch the tube wall and bottom. Ad...
Embodiment 3
[0066] Example 3 reverse transcription of miR-21 and miR-16
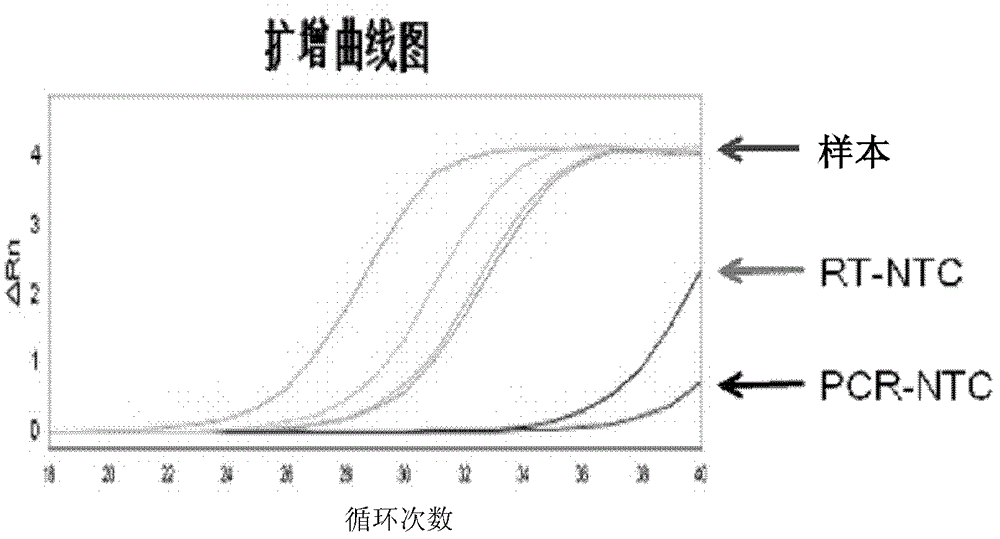
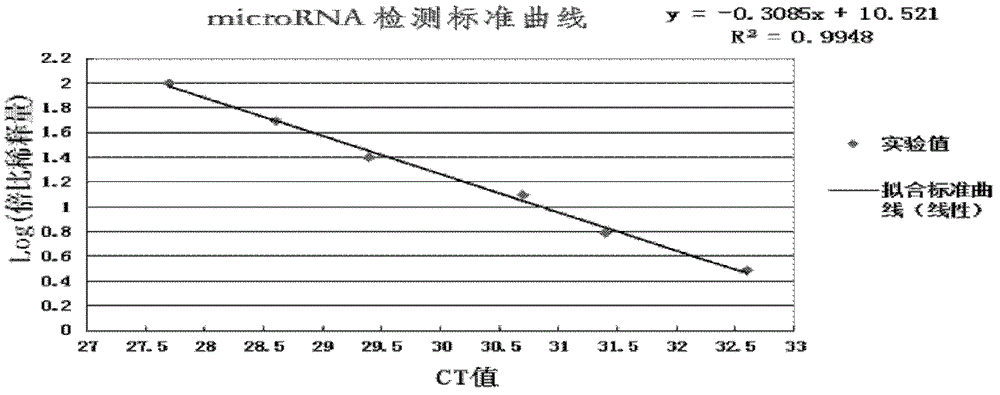
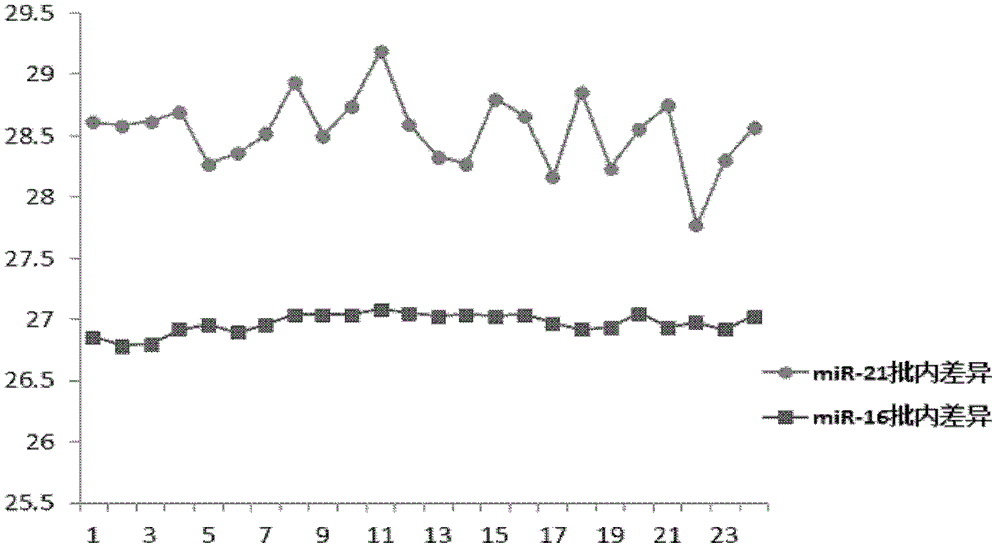
[0067] The RNA extracted in Example 2 was reverse-transcribed with a self-assembled reverse transcription kit. Add 23 μL of extracted RNA and 23 μL of DEPC water [as a reverse transcription no-template control (RT-NTC)] to various reagents in the reverse transcription kit, which contain a specific volume of 500 nmol / L miR-21 or miR-16 stem-loop Reverse transcription primer 1μL, 200U / μL M-MLV 1μL, reverse transcription buffer 2μL, 1mol / 1DTT 0.1μL, 10mmol / 1DNTPs 0.5μL and 40U / μL RNAsin 0.1μL. The reaction conditions were: annealing at 16°C for 30 minutes, reverse transcription at 42°C for 60 minutes, and reverse enzyme inactivation at 85°C for 5 minutes. Then, the reverse transcription product was stored at -20°C.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com