Novel heatproof beta-agaropectinase AgaXa, its preparation method and application
A technology of agarase, bl21-pet32-agaxa, applied in the direction of DNA preparation, botany equipment and methods, biochemical equipment and methods, etc., can solve hydrolyzate easily destroyed, poor specificity, agar oligosaccharide reaction serious problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Example 1: Cloning of β-agarase AgaXa
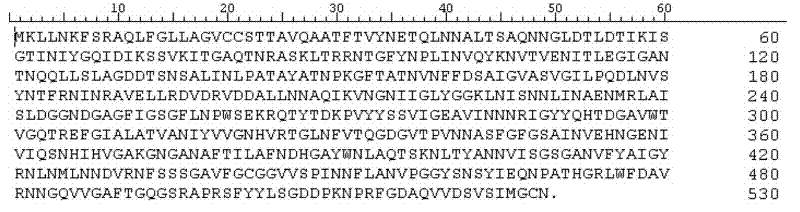
[0021] pass Eco RI and Hin dⅢ These two enzymes double digest Catenivulum For the total DNA of sp. X3, recover the gene fragment with a length of 3-7kb after digesting the bacterial genome, and connect it with the pBluescriptⅡSK(+) vector that has undergone the same digestion, to construct a partial genome library of the bacteria. Screen the positive clones with agarase activity on the plate and send them to Beijing Huada Company for sequencing. The complete open reading frame of a gene was analyzed by the biological software DNASTAR. The forward primer (5'-GCGCGGATCCATCAAACTATTGAAT-3') and the reverse primer (5'-GGCCTCGAG AATTACAACCCAT-3') were designed by the biological software Primer5.0, and the genomic DNA was used as a template for PCR.
[0022] Reaction system (25 μL): 1 μL DNA template, 1 μL forward primer, 1 μL reverse primer, 9.5 μL double distilled water, 12.5 μL high-fidelity PCR mix.
[0023] Reaction conditi...
Embodiment 2
[0026] Example 2: Construction of the Escherichia coli expression vector pET32-agaXa, the β-agarase gene agaXa obtained in Example 1 and the expression vector pET32a (+) were respectively used Bam HI and xho Ⅰ The conditions for double enzyme digestion are as follows:
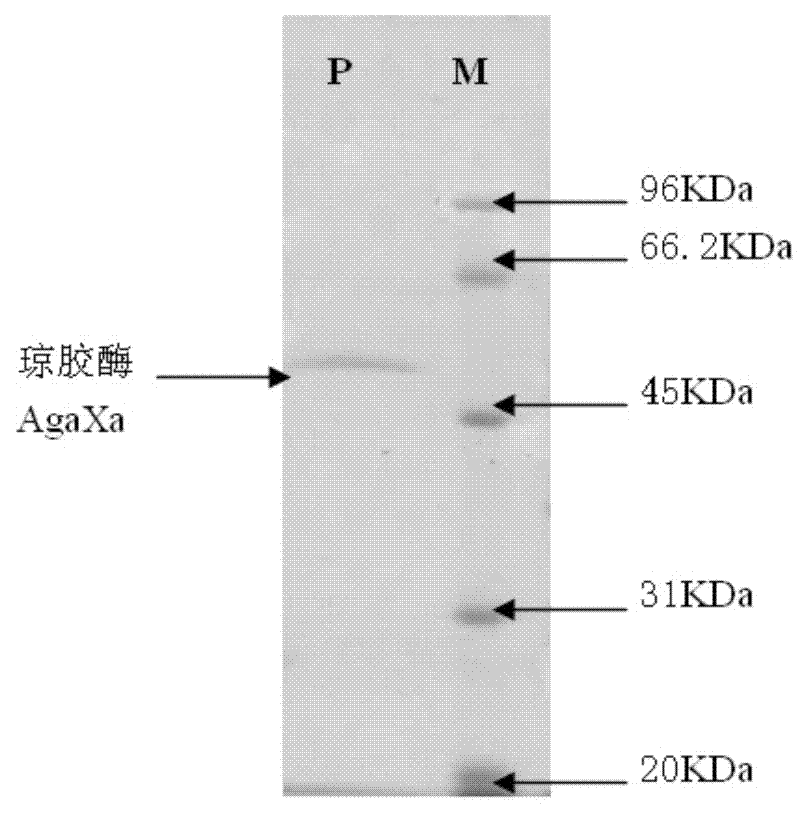
[0027] Reaction system (20 μL): double distilled water 10 μL, 10×K Buffer 2 μL, gene agaXa DNA [pET32a (+) plasmid DNA] 6 μL, Bam H 1 μL, xho Ⅰ1 μL, digestion at 37°C for 2 hours. After electrophoresis, target fragments with sizes of about 1.5kb and 5.9kb were recovered respectively. The two are β-agarase gene agaXa and expression vector pET32a (+), respectively.
[0028] then use T 4 The ligase connects the two, and the reaction conditions are as follows:
[0029] Reaction system (20μL): 9μL of double distilled water, 2μL of 1.5kb fragment, 6μL of 5.9kb fragment, 10×T 4 Ligase Buffer 2μL, T 4 Ligase 1 μL, at 16°C for more than 8 hours to obtain the Escherichia coli expression vector pET32-agaXa.
Embodiment 3
[0030] Example 3: Construction of recombinant Escherichia coli BL21-pET32-agaXa.
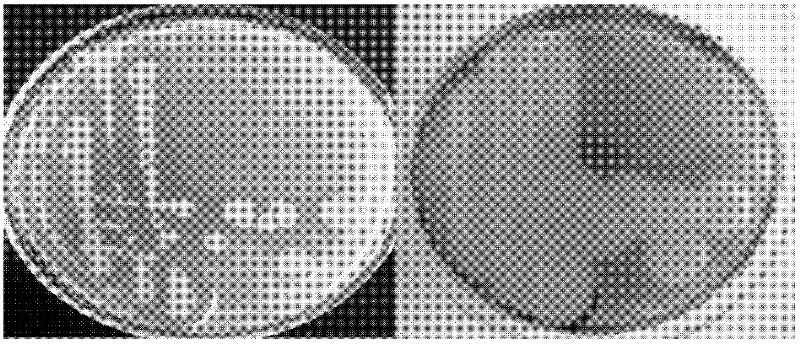
[0031] Take 10 μL of the recombinant vector pET32-agaXa obtained in Example 2 and add it to 100 μL BL21 competent cells, and place on ice for 30 minutes; then heat shock at 42°C for 90 seconds; quickly transfer the EP tube to the ice bath for 15 minutes; Add 700 μL of LB medium to the tube, transfer the tube to a shaker at 37°C, and shake slowly for 60 minutes to recover the bacteria; take 50 μL of the recovered bacteria solution and spread it on a solid LB plate containing Amp, X-Gal and IPTG; invert the plate , incubate at 37°C for 24 hours; finally pick a single white colony for PCR detection. In agarose electrophoresis, the specific band with a size of 1.5kb is the Escherichia coli recombinant BL21-pET32-agaXa containing pET32-agaXa ( image 3 ).
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com