Taq DNA (deoxyribonucleic acid) polymerase cold start activity detection metho
A technology for activity detection and polymerase, which is applied in biochemical equipment and methods, microbial measurement/inspection, fluorescence/phosphorescence, etc. It can solve the problems of poor specificity of PCR products, base mismatches, and high temperature resistance of Klenow fragments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
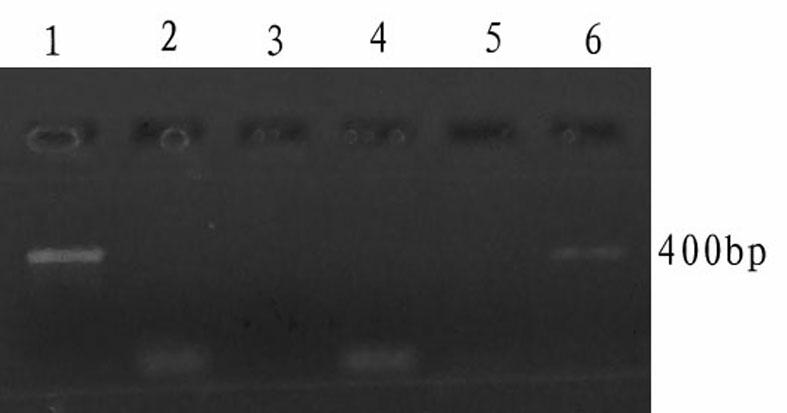
[0032] Use the plasmid pGEM-5Zf(+) as a template to detect the DNA polymerase cold-start activity of the Taq DNA polymerase to be tested. The specific steps are as follows:
[0033] (1) Design and synthesis of primers and pseudo-templates
[0034] Referring to the complete gene sequence of the plasmid pGEM-5Zf (+) registered on GenBank, a pair of primers were designed using PP5.0 software. Artificially synthesized, the theoretically amplified sequence length of this pair of primers is 401bp, and its base sequence is as shown in SEQ ID NO:4. Simultaneously design the false template T3, the sequence is as follows: T3: 5'-AAGCAGTTCACTAGAGGTATGTAGGCGGTGCTA -3', wherein, the 3' end of T3 is completely complementary to T2, and the 13 bases at the 5' end are compatible with the template plasmid pGEM-5Zf (+) The sequence is completely different.
[0035] (2) Treatment of false templates and primers before PCR reaction
[0036] Dilute the synthesized primers T1 and T2 and the false ...
Embodiment 2
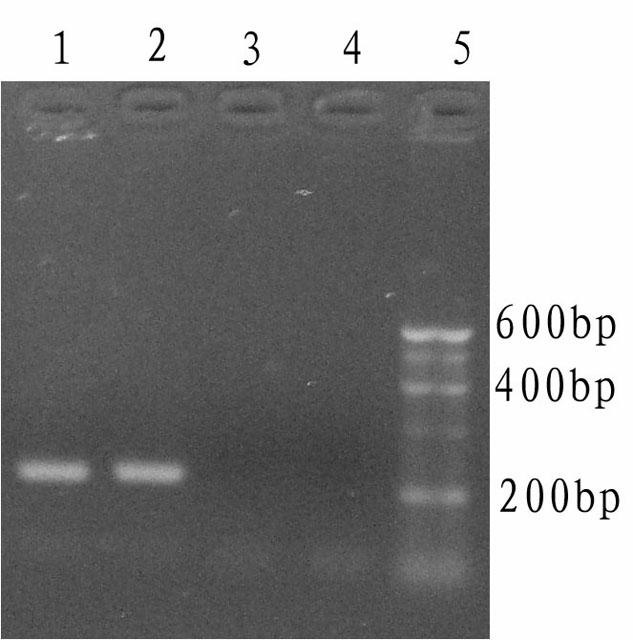
[0048] The DNA polymerase cold-start activity detection of the Tth DNA polymerase to be tested is performed using the whole human gene as a template, and the specific operation steps are as follows:
[0049] (1) Design and synthesis of primers and pseudo-templates
[0050] Referring to the complete human HLA-DRA1 gene sequence registered on GenBank, a pair of primers were designed using the primer design software primer5.0. Synthesized, the theoretically amplified sequence length of this pair of primers is 223bp, and its base sequence is shown in SEQ ID NO: 8; at the same time, a false template T6 is designed, and the sequence is as follows: T6: 5'-TACTAGCGCGTG GTTTGACTTTGATGGTGATGAGA-3' Among them, the 3' end of T6 It is completely complementary to T5, and the 12 bases at the 5' end are completely different from the corresponding sequence of the HLA-DRA1 gene of the template human genome, and are artificially synthesized;
[0051] (2) Treatment of false templates and primers...
Embodiment 3
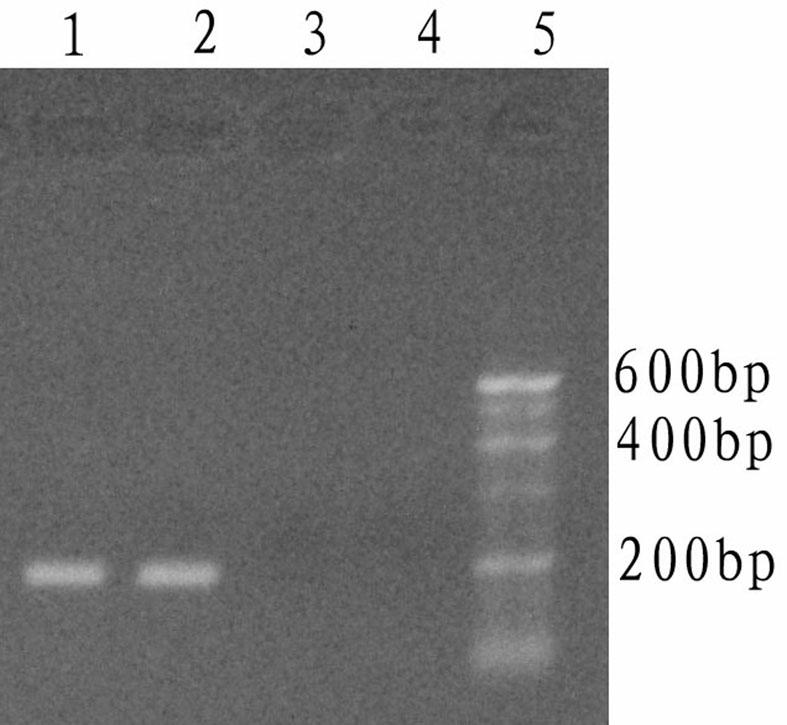
[0064] The DNA polymerase cold-start activity detection of the Taq DNA polymerase to be tested is performed using the whole mouse gene as a template. The specific operation steps are as follows:
[0065] (1) Design and processing of primers and pseudo-template sequences
[0066] Referring to the complete gene sequence of mouse GAPDH (glyceraldehyde-3-phosphate dehydrogenase) registered on GenBank, a pair of primers were designed using the primer design software primer5.0. The sequence is as follows: T7: 5'- CCCCTGTTTCTTGTCTTTCA-3', T8: 5'- GCTTCCCATTCTCGGCCTTG-3' is artificially synthesized, and the theoretically amplified sequence length of this pair of primers is 191bp, and its base sequence is as shown in SEQ ID NO:12. Simultaneously design a false template T9, the sequence is as follows: T9: 5'- CGAATGTGAC CAAGGCCGAGAATGGGAAGC-3', wherein, the 3' end of T9 is completely complementary to T8, and the 10 bases at the 5' end are completely complementary to the sequence of the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com