Method for knocking out gene in clostridium acetobutylicum
A clostridia and gene technology, applied in the field of genetic engineering, can solve the problems of low homologous recombination efficiency, difficulty in obtaining double-crossover transformants, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0097] Embodiment 1, the construction of pUC19-adc plasmid vector
[0098] 1. Construction of pUC19-adc plasmid vector
[0099] Using the Clostridium acetobutylicum genome as a template, primers HindIII (adc-L1) and R1-L1 (adc) were used to amplify adc upstream homology arm adcL by PCR; primers L2-R2 (adc) and BamHI (adc- R2) Amplify the downstream homology arm adcR of adc by PCR; after the PCR product is recovered (see the Huashun Glue Recovery Kit for the method), it is digested with BamHI and HindIII and ligated into the same digested plasmid pUC19 (see the Huasun Glue Recovery Kit for the method) ; Use T4 ligase (Takara Company) to ligate the digested fragment and the vector, and then overnight at 16°C. The resulting plasmid was named pUC19-adc.
[0100] Use pUC19-adc to transform E.coli DH5α competent cells, see Molecular Cloning 3 for the method. Then the recombinants were picked, cultured in LB test tubes (Amp) at 37°C overnight, the plasmids were extracted, and iden...
Embodiment 2
[0109] Embodiment 2, the construction of pSY9-1-adc plasmid vector
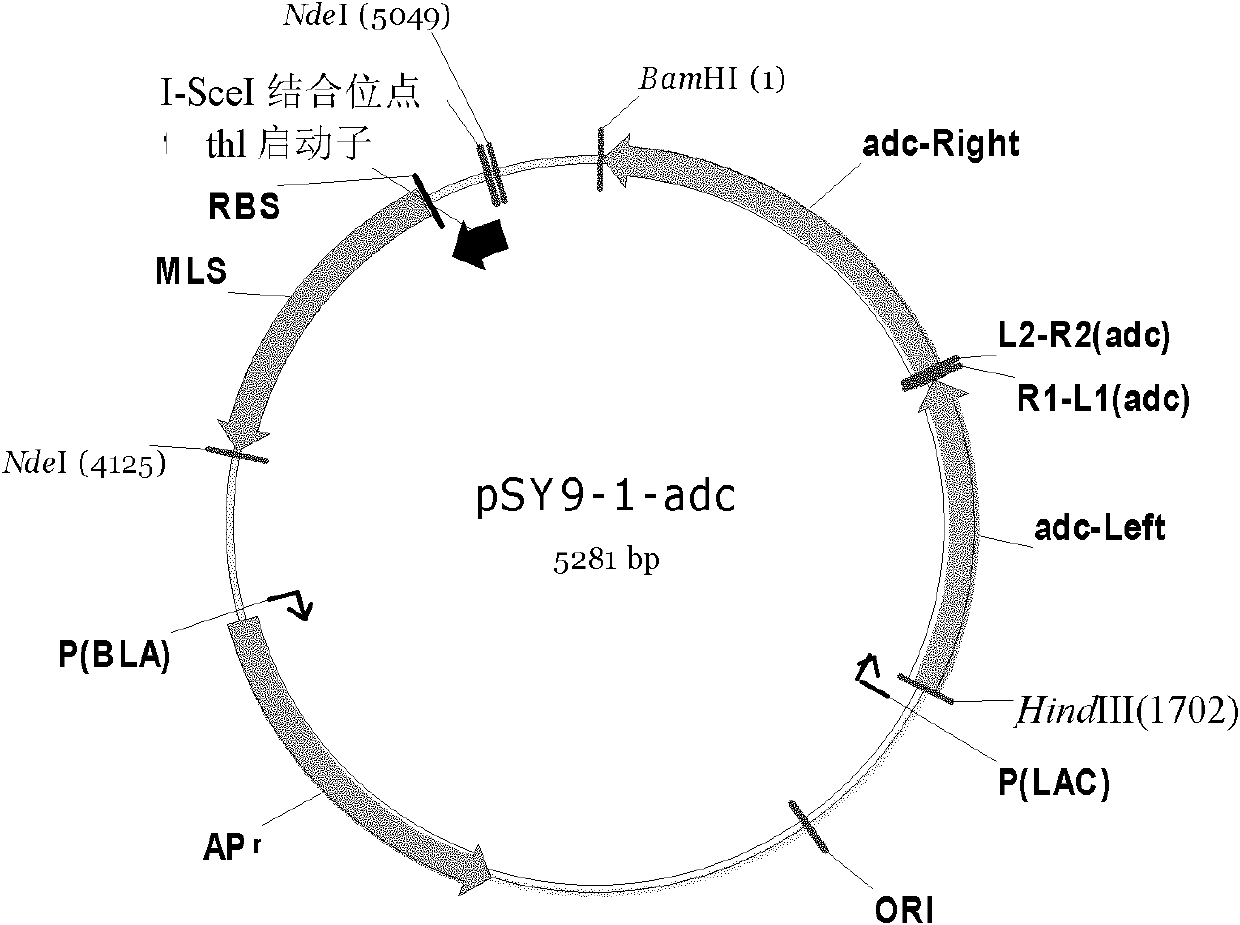
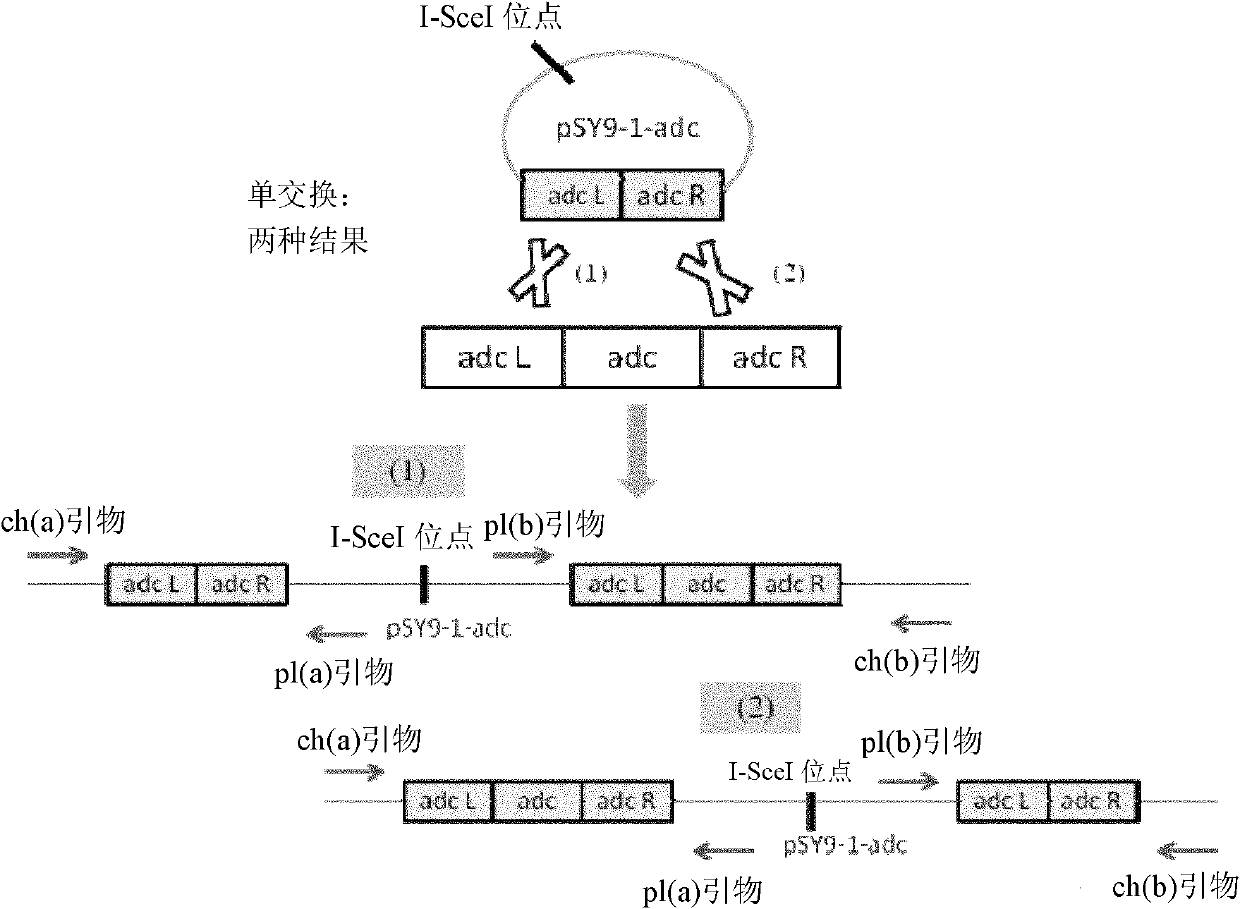
[0110] Using the Clostridium acetobutylicum genome as a template, primers NdeI (SceI-thl1), Eml-Thl1 were used to amplify the thl promoter (including the rbs sequence) by PCR, and introduced the I-SceI homing enzyme recognition site 18bp; at the same time Using the pIMP1 plasmid as a template, using Thl2-Em2 and NdeI (XholI-Em-2) as primers, PCR amplifies the Em resistance gene, respectively purifying the PCR products, using it as a template, and using NdeI (SceI-thl1), NdeI (SceI-thl1), NdeI (XholI-Em-2) was used as a primer, and the sequence containing thl promoter + RBS + MLS was obtained by Overlapping PCR. After digestion with NdeI, it was connected into the pUC19-adc plasmid with the same restriction enzyme digestion. Enzyme digestion identification chart see Figure 13 ), select the forward transformant, named as pSY9-1-adc (see figure 1 and SEQ ID NO: 2).
[0111] The primer sequences used are as f...
Embodiment 3
[0117] Embodiment 3, the construction of pSY14-thl plasmid vector
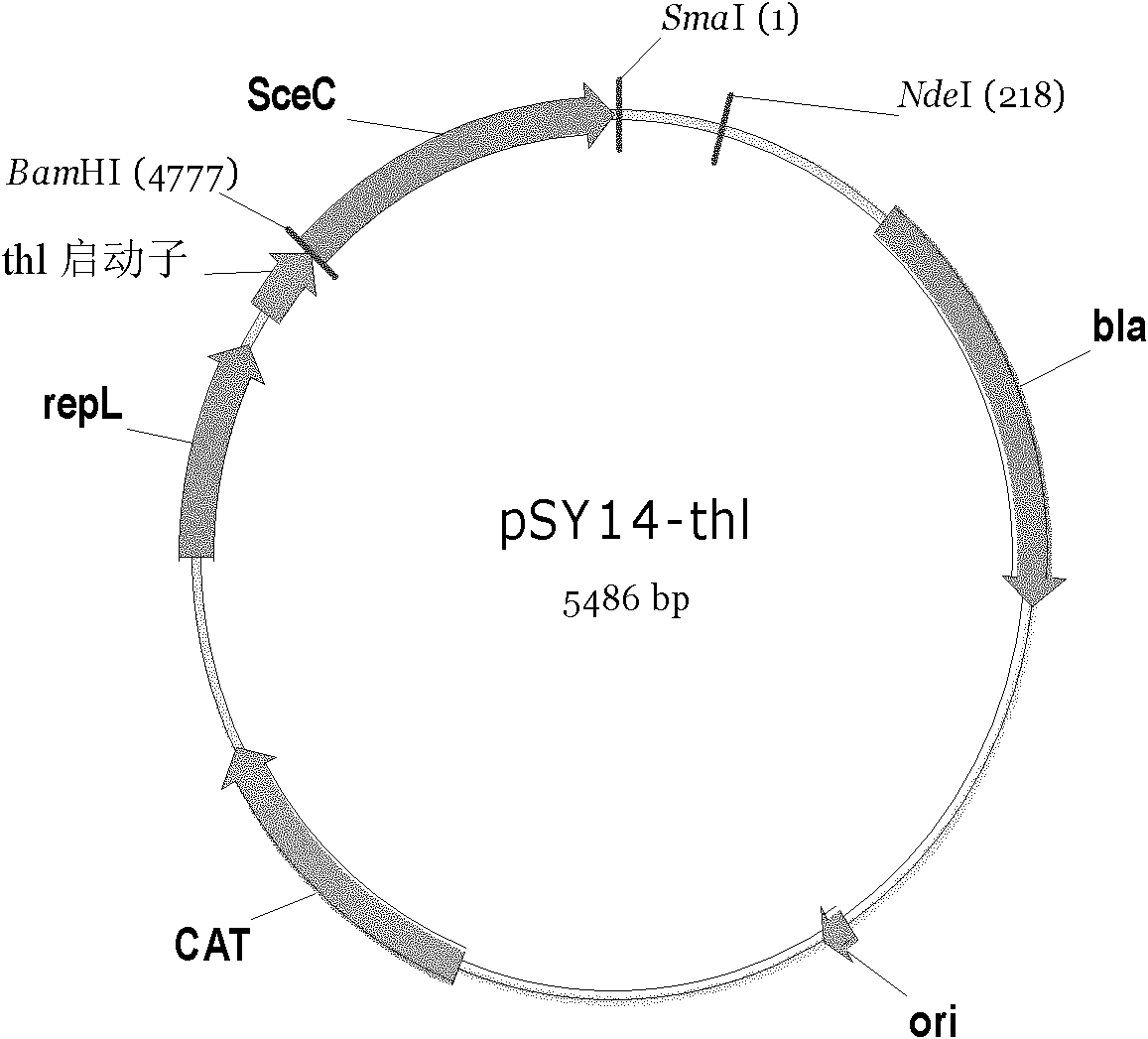
[0118] Due to the large gap between the original sequence of I-SceI and the use of codons in Clostridium, the inventors optimized the codons of the gene, which was synthesized by Nanjing Jinsiruiji Biotechnology Co., Ltd., and the synthesized gene was cloned in a commercially available The pUC57 plasmid is named: pUC57-SceC. The sceC sequence (SEQ ID NO: 1) was amplified by PCR with primer SceC-1 (BamHI) and primer SceC-2 (SmaI). After double digestion with BamHI and SmaI, it was connected into the pITC plasmid digested with the same enzyme. After enzyme digestion verification (for the enzyme digestion identification diagram, see Figure 14 ), named pSY14-thl (see figure 2 and SEQ ID NO: 3).
[0119] The primer sequences used are as follows:
[0120] SceC-1 (BamHI): 5`-CGCGGATCCATGCATCAAAAGAATCAAGT-3` (SEQ ID NO: 12)
[0121] SceC-2 (SmaI): 5`-TCCCCCGGGTTATTTTAAGAAAAGTTTCAC-3` (SEQ ID NO: 13)
[0122] Oth...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com