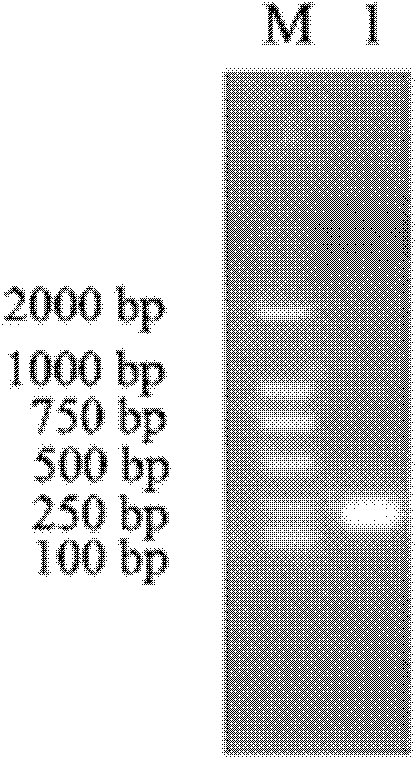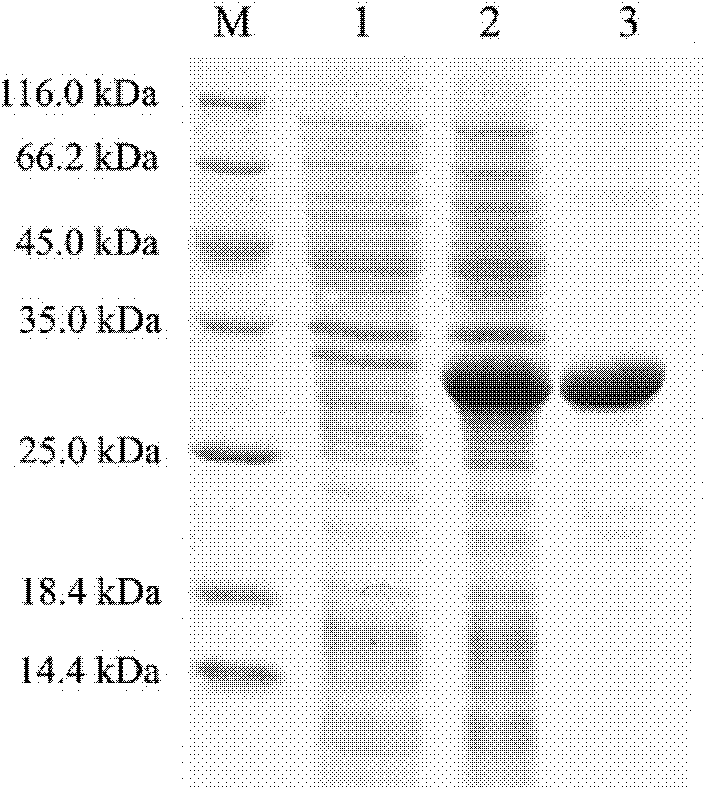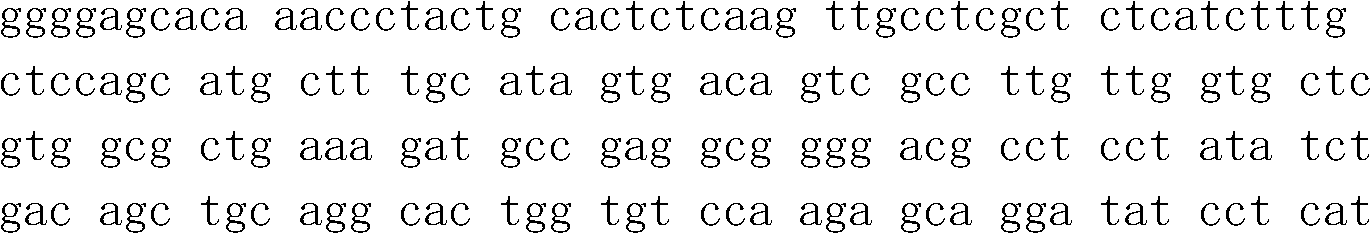Portunus trituberculatus PtCrustin-2 gene, and coded protein and application thereof
A technology of swimming crab and gene encoding, which is applied in the field of molecular biology and can solve the problems that the research on the crustin of the swimming crab is less and other problems.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] The PtCrustin-2 gene of Portunus trituratus is the base sequence shown in SEQ ID No.1.
[0026] Sequence Listing SEQ ID No.1 is:
[0027]
[0028]
[0029] (a) Sequence features
[0030] ●Length: 629bp (effective length 58-405)
[0031] ●Type: base sequence
[0032] ●Chain type: single chain
[0033] ●Topology: Linear
[0034] (b) Molecular type: double-stranded DNA
[0035] (c) Assumption: No
[0036] (d) Antonym: No
[0037] (e) Original source: Portunus trituberculatus
[0038] (f) Distinctive name: CDS
[0039] The specific operation of the construction is as follows:
[0040] 1. Extraction of total RNA and purification of mRNA from Pool crab: Trizol reagent from Invitrogen company was used to extract total RNA from pool crab, and mRNA was purified by Oligitex mRNA purification kit from QIAGENE company.
[0041] 2. Construction of the cDNA library of Portunus trituratus: The cDNA library was constructed using the instructions of the Creator Smart cDN...
Embodiment 2
[0059] The base sequence of the PtCrustin-2 sequence table of PtCrustin-2 is described in SEQ ID No. 1 of the sequence table, and the amino acid sequence of the amino acid sequence is described in the SEQ ID No. 2 of the sequence table.
[0060] Sequence Listing SEQ ID No.2 is:
[0061]
[0062]
[0063] It has a complete encoded protein containing 98 amino acids, of which 1-20 amino acids of the coding sequence are signal peptides, and the mature peptide contains 95 amino acids, the predicted molecular weight is 10.58kDa, and the isoelectric point is 6.74. The mature peptide has a typical type I Crustin pattern structure, including a cysteine-rich region (Cys-rich region) and a WAP domain (WAP domain). Among them, the arrangement of 4 cysteine residues in the cysteine-rich region is relatively conservative, in a C-(X3)-C-(X8-12)-C-C pattern; the WAP domain contains 8 cysteine residues acid residues, which are capable of forming four disulfide cores (4-DSC).
[006...
Embodiment 3
[0067] In vitro antibacterial test of PtCrustin-2 recombinant protein from Portunus trituratus:
[0068] 1. Culture and preparation of microorganisms: TSB medium for Vibrio alginolyticus at 28°C, TSB medium for Pseudomonas aeruginosa at 37°C, LB medium for Staphylococcus aureus at 37°C, and LB medium for Micrococcus luteus 37°C, Pichia pastoris was incubated with YPD medium at 28°C, and each of the above strains was incubated with a shaker at 220 rpm / min so that the bacterial concentration reached the logarithmic growth phase, the cells were diluted with 50 mM Tris-HCl (pH=8.0) buffer solution, The number of colonies per milliliter of bacterial solution is about 1 × 10, respectively. 3 .
[0069] 2. Determination of the antibacterial activity of the recombinant protein PtCrustin-2: The recombinant protein PtCrustin-2 obtained in the above example was diluted with Tris-HCl (50mM, pH=8.0) in a gradient (1, 1 / 2, 1 / 4, 1 / 8, After 1 / 16, 1 / 32, 1 / 64, 1 / 128), 50 μl was added to a ste...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com