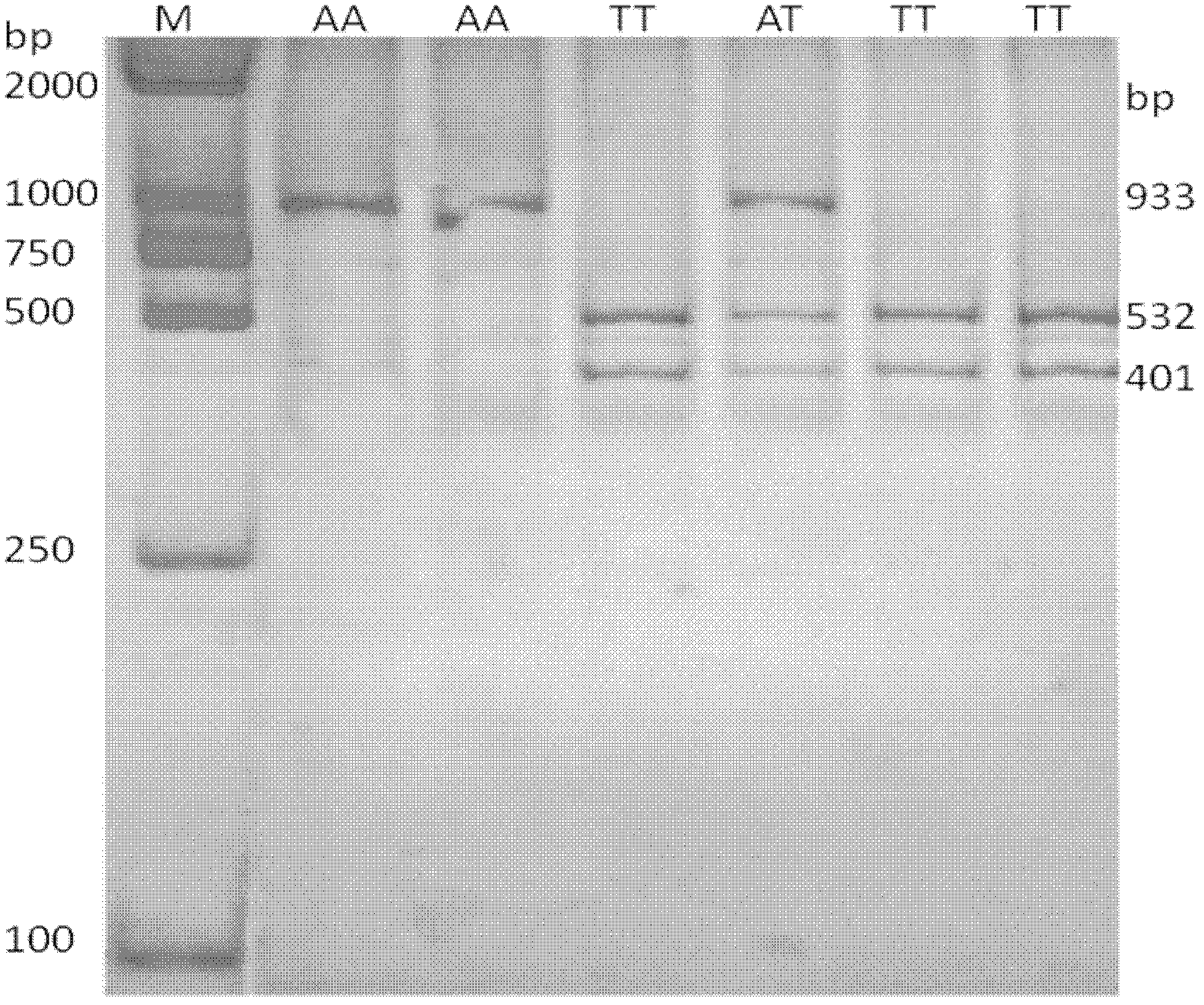Method of screening SNP (single nucleotide polymorphism) sites in HSTN (histatherin) gene of cow with mastitis resistance and kit
A mastitis and kit technology, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., to reduce detection costs and improve detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
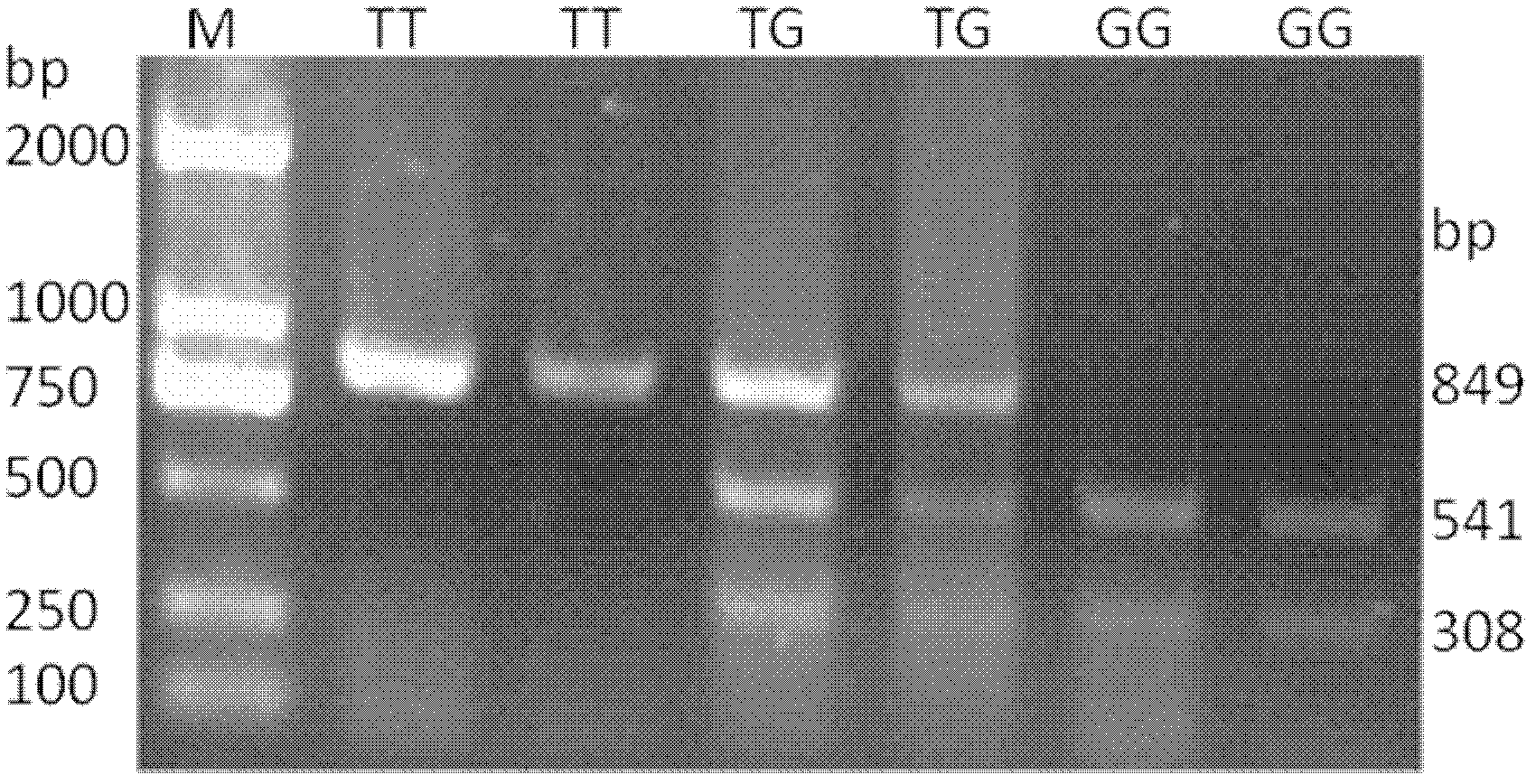
[0043] Example 1 Haplotype combination and determination of mutation sites
[0044] The present invention adopts PCR sequencing and PCR-RFLP technology to screen the mutation of dairy cattle HSTN gene, and determines three mutation sites of 635, 1607 and 12624 through enzyme-cut electrophoresis bands and comparison of sequencing results. Haplotypes were constructed using Shesis software.
[0045] 1.1 Collection of milk cow jugular vein blood (the Holstein cow jugular vein blood used in the present invention is taken from large-scale cattle farms in Shandong Province, including Jinan Jiabao, Qingdao Yimu, etc., which are the direct source and original source of the blood sample used in the present invention), ACD anticoagulation, using the conventional phenol / chloroform extraction method to extract genomic DNA, DNA samples were diluted to 50ng / μL for later use. A total of 500 experimental cows came from large-scale dairy farms in Shandong Province.
[0046] 1.2 Primer design ...
Embodiment 2
[0062] Example 2 Correlation between HSTN Gene Haplotype Combination and Dairy Cow Mastitis Resistance
[0063] Determination of somatic cell score: Collect 40 mL of milk sample, add 0.03 g of potassium dichromate, mix gently, and then send it to the DHI laboratory of the Dairy Center of Shandong Academy of Agricultural Sciences to use the Foss 6000 milk component and somatic cell online analysis system (Foss MilkScan FT 6000, Fossmatic 5000) to measure the number of somatic cells. According to the formula SCS=log2(SCC / 100)+3 (wherein SCS-somatic cell score; SCC-somatic cell number, unit thousand / mL), the individual's somatic cell score was calculated.
[0064]The least square analysis of variance adopts the ANOVA-Linear Models program of SAS 9.0 (Statistical Analysis System) software to establish the following model, and perform the least square analysis and significance test on the somatic score and haplotype. The model is:
[0065] Y ijkl =μ+G i +E j +S k +P l +e ijk...
Embodiment 3
[0072] Example 3 Detection Kit
[0073] A kit for screening dairy cows based on HSTN gene haplotype combination markers, as described in Example 1, the haplotypes constructed by the 635 A→T mutation, 1607 T→C and 12624 T→G mutations in NC_007304.5 Combinations were strongly associated with mastitis resistance traits in dairy cows. Therefore, HSTN gene-specific primers can be designed based on these three SNP sites for amplification and detection.
[0074] Prepare the kit (100 times) for detecting mastitis resistance traits of dairy cows, the composition is as shown in Table 4:
[0075] Table 4
[0076]
[0077] The bovine blood was collected, the total DNA was extracted, and the reaction was carried out according to the method described in Example 1. Dilute the PCR primers in the kit to 10 μmol / L, and use the extracted DNA as a template to carry out PCR reaction with the above kit. PCR reaction conditions: Denaturation at 95°C for 5min, denaturation at 94°C for 30s, ann...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com