Method for detecting miRNA (micro ribonucleic acid) based on graphene/nucleic acid dye platform
A nucleic acid dye and graphene technology, which is applied in the field of nucleic acid fluorescence detection, can solve the problems of cumbersome operation, low sensitivity, and time-consuming, and achieve the effects of simple operation, common detection devices, and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
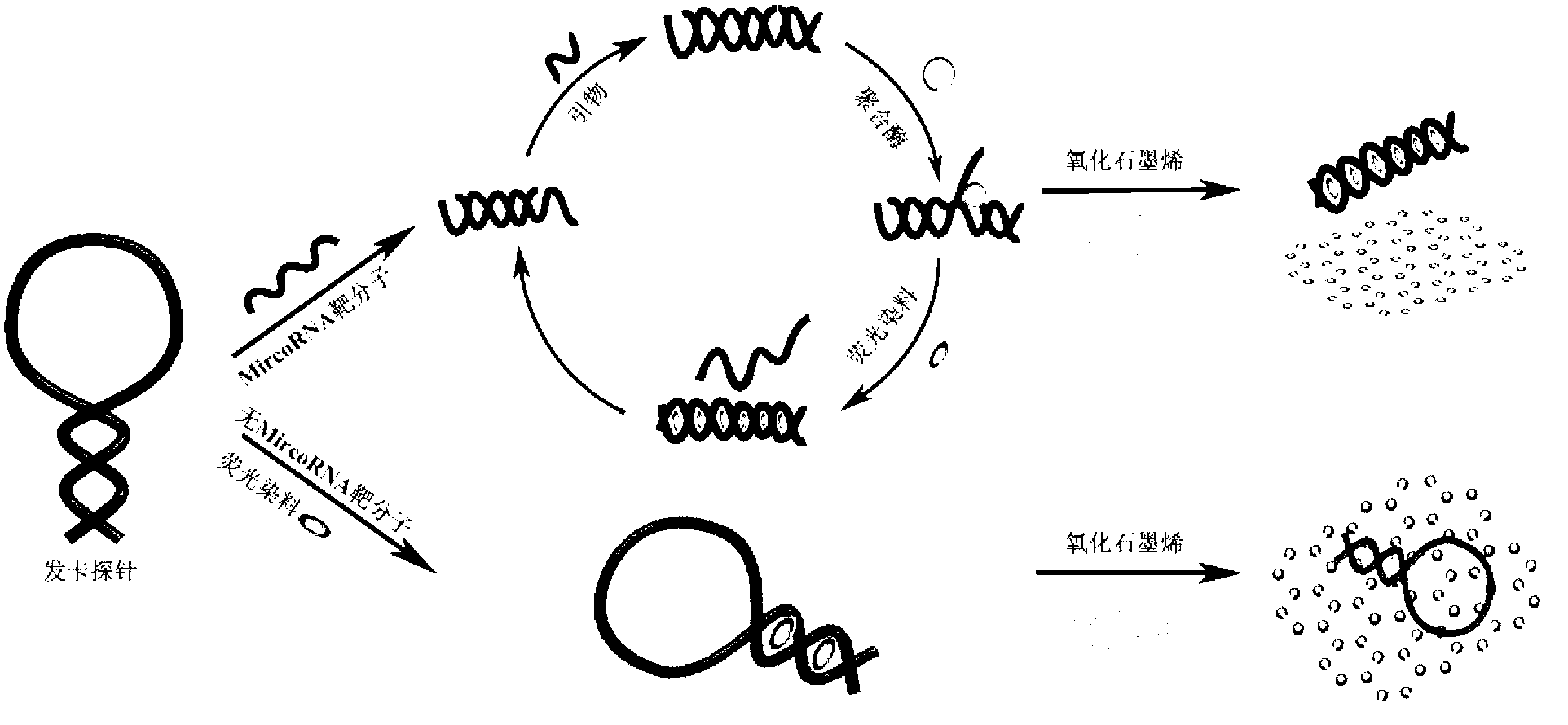
[0052] The construction of embodiment 1 graphene oxide / SYBR Green I detection platform
[0053] (1) Design a DNA hairpin probe for miR-21 as the target molecule, and the binding subsequence of the DNA hairpin probe is TT with both 5' and 3' ends;
[0054] (2) Take 1 mL of graphene oxide with a diameter of 1 μm (Pioneer Nanomaterials Co., Ltd.), wash it twice with double distilled water at room temperature, and centrifuge it for later use;
[0055] (3) Add the designed DNA hairpin probe, graphene oxide, SYBR Green I, different concentrations of miR-21 target molecules and detection buffer to a 96-well plate for hybridization reaction; reaction specific parameters For: 50nM DNA hairpin probe, 8μg / mL graphene oxide, 2μM SYBR Green I nucleic acid dye (invitrogen company), Tris-HCl 10mM and MgCl in 50μL buffer (pH 8.2) reaction system 2 5mM, hybridization at 37°C for 30 minutes; the concentrations of miR-21 target molecules in the five samples were 0nM, 2nM, 10nM, 50nM, and 250nM;...
Embodiment 2
[0057] Example 2 Strand Displacement Isothermal Amplification Reaction
[0058] (1) Prepare strand displacement constant temperature amplification reaction system: 20 μL reaction system contains 200 nM DNA hairpin probe, 400 nM primers, 2U Klenow fragment exo-polymerase (MBI company), Klenow fragment exo-polymerase buffer, dNTPs (each 0.1mM), volume ratio of 3% DMSO and 50fmol target miR-21;
[0059] (2) The reaction system in step (1) is reacted at 37°C for 30-60 minutes;
[0060] The result of the strand displacement isothermal amplification reaction is as follows: image 3 Shown: 1. DNA hairpin probe, 2. DNA hairpin probe and primers, 3. DNA hairpin probe, primers and strand displacement constant temperature amplification enzyme, 4. DNA hairpin probe, target miR-21 and primers, 5 .DNA hairpin probe, target miR-21, primers and strand displacement constant temperature amplification enzyme (react at 37°C for 30 minutes), 6. Same as 5 (react at 37°C for 60 minutes); In the p...
Embodiment 3
[0061] Example 3 Detection of miR-21 using graphene oxide / SYBR Green I detection platform combined with strand displacement constant temperature amplification technology
[0062] (1) Design DNA hairpin probes and primers according to the sequence for detecting miR-21 target molecules;
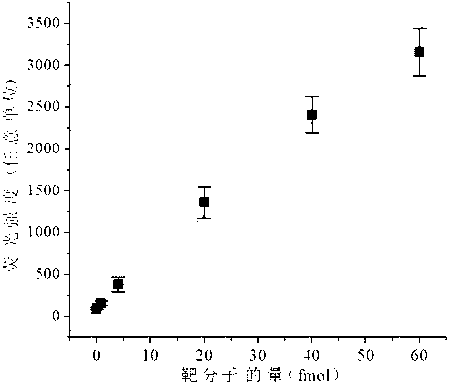
[0063] (2) DNA hairpin probes, primers, different amounts of miR-21 target molecules, DMSO, enzymes with strand displacement amplification properties and necessary reactants (Klenow fragment exo-polymerase from MBI company and its standard Buffer, dNTPs) were mixed for strand displacement constant temperature amplification reaction; the specific parameters of the reaction were: 20μL reaction system containing 200nM hairpin probe, 400nM primers, 2U Klenow fragment exo-polymerase, dNTPs (each 0.1mM), volume The ratio of 3% DMSO and the amount of miR-21 target molecules are 0.4fmol, 0.8fmol, 4fmol, 20fmol, 40fmol, 60fmol, 100fmol respectively, and react at 37°C for 60 minutes;
[0064] (3) Add 60...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com