Method utilizing degenerate primer to detect diversity of bacteria catalase in seawater
A technology of bacterial catalase and catalase, which is applied in the direction of biochemical equipment and methods, and the determination/inspection of microorganisms, can solve the problem of time-consuming, expensive, and undeveloped catalase diversity detection methods and other problems, to achieve the effect of convenient use and precise method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] 1) Determination of degenerate primers:
[0031] Collect various known bacterial catalase protein sequences from GenBank and perform multiple sequence comparisons to find out the conservative amino acid sequences, thereby obtaining suitable degenerate primers for PCR amplification of catalase genes. There are four kinds of forward primers and two reverse primers. The sequence of forward primers (5' to 3') is:
[0032] C3+: TTTYAAYCGAGARMGRGTNCCNGARMGNG;
[0033] CA: GTACCTGAAMGRGTRGTSCAYGCNMRRGG;
[0034] The reverse primer sequence (5’ to 3’) is:
[0035] CB: GAATATGCAAAAAGGCGNCCYTGNARNAKYTTRTC;
[0036] C3-:AATCTTTATGAGGCCAAACTTTTGTNANRTCRAA. ,
[0037] 2) Use forward degenerate primer CA:CCNGARMGNGTNGTNCAYGCN and reverse degenerate primer C3-:
[0038] AATCTTTATGAGGCCAAACTTTTGTNANRTCRAA, PCR amplification of catalase gene fragment. PCR amplification uses 12.5 μl reaction system: 2.5 mM dNTP 1 μl, 10×Taq PCR amplification buffer 1.25 μl, 50 μM two degenerate primers each 0.15 μ...
Embodiment 2
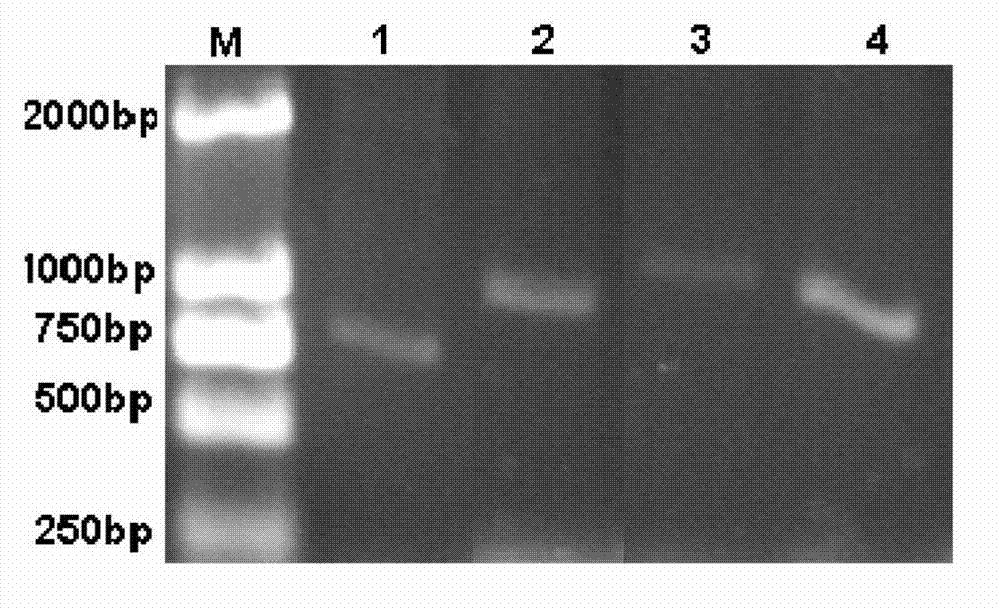
[0043] 1) Use forward degenerate primer CA:CCNGARMGNGTNGTNCAYGCN and reverse degenerate primer CB:AAATGTTCGGCCYTGNARNADYTTRTC to amplify catalase gene fragments by PCR. PCR uses 12.5 μl reaction system: 2.5 mM dNTP 1 μl, 10×Taq PCR buffer 1.25 μl, 50 μM two degenerate primers each 0.15 μl, Taq DNA polymerase 0.625U, seawater DNA 1 μl, Add water to a total volume of 12.5 microliters. Six 12.5 microliter PCR systems were used to prepare each catalase gene fragment. PCR conditions: 94℃2min; 94℃30s, 60℃→51℃1min, 72℃1min 30s, 10 cycles, each cycle reduces the annealing temperature by 1℃; 94℃30s, 50℃1min, 72℃1min, 20 cycles; 72 ℃ 3min. After the reaction, 6 tubes of PCR products were combined, and a single PCR product with a molecular weight of 500bp to 1000bp was purified using a gel recovery kit.
[0044] 2) Use the T vector kit to connect the PCR products of step 1) and transform E. coli competent cells to construct a T vector library of catalase gene fragments. Blue-white scree...
Embodiment 3
[0048] 1) Use forward degenerate primer C3+:TTTYAAYCGAGARMGRGTNCCNGA and reverse degenerate primer CB:AAATGTTCGGCCYTGNARNADYTTRTC to amplify catalase gene fragments. PCR uses 12.5 μl reaction system: 2.5 mM dNTP 1 μl, 10×Taq PCR buffer 1.25 μl, 50 μM two degenerate primers each 0.15 μl, Taq DNA polymerase 0.625U, seawater DNA 1 μl, Add water to a total volume of 12.5 microliters. Six 12.5 microliter PCR systems were used to prepare each catalase gene fragment. PCR conditions: 94℃2min; 94℃30s, 60℃→51℃1min, 72℃1min 30s, 10 cycles, each cycle reduces the annealing temperature by 1℃; 94℃30s, 50℃1min, 72℃1min, 20 cycles; 72 ℃ 3min. After the reaction, 6 tubes of PCR products were combined, and a single PCR product with a molecular weight of 500 bp to 1000 bp was purified using a gel recovery kit.
[0049] 2) Use the T vector kit to connect the PCR products of step 1) and transform E. coli competent cells to construct a T vector library of catalase gene fragments. Blue-white screen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com