High selectivity gene mutation detection method, high selectivity gene mutation detection primer and primer design method
A technology with high selectivity and detection method, applied in DNA/RNA fragments, DNA preparation, recombinant DNA technology, etc., can solve the problems of high cost, difficult synthesis of MGB probes, unfavorable wide application, etc., and achieve high selectivity, Low cost, high sensitivity effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
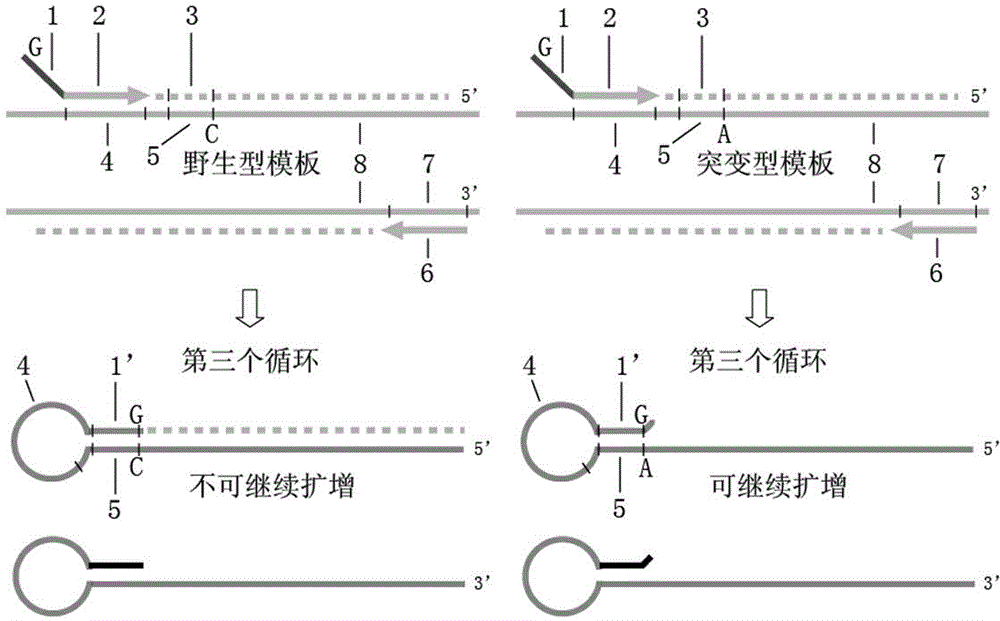
[0027] Select SNP (rs13182883) as the research object, its genotype A is the wild type, and the genotype G is the mutant type, and the corresponding primers and probes are designed as follows:
[0028] Primer F: TGAGGATTTG CTGGCCTGCAGTGAGCATT
[0029] Primer R: CTGACAATCCTTGGTCTGCT
[0030] Detection probe: FAM-GAAGAAAGTAACGGAGAGCCTG-BHQ
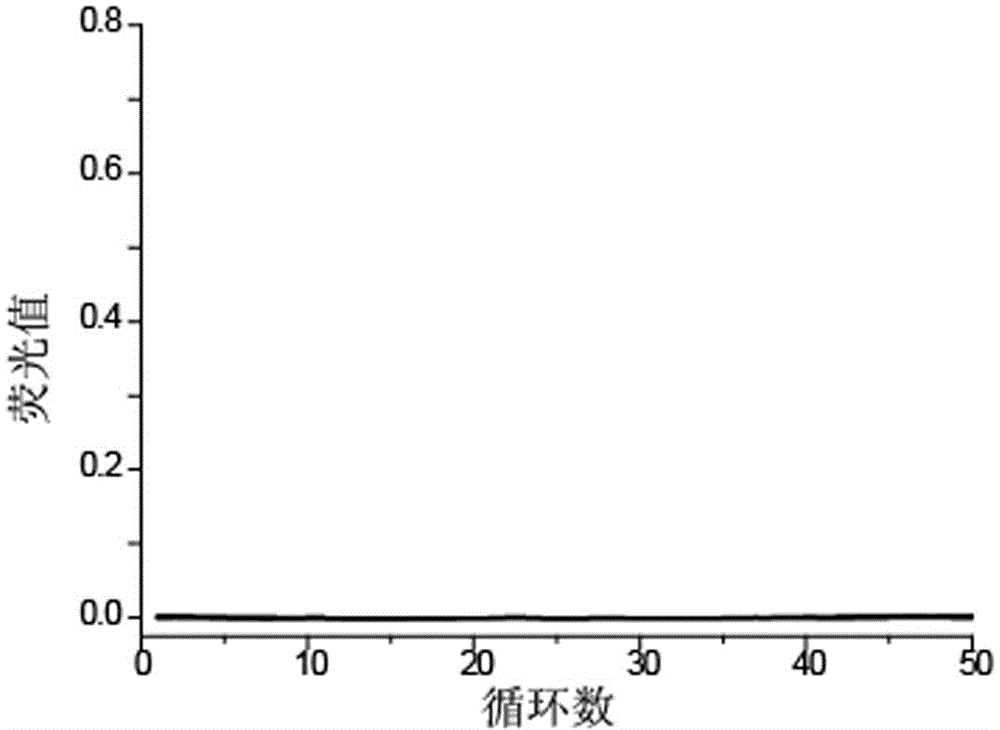
[0031] In order to investigate the specificity of the method, homozygous wild template 1 was serially diluted 10 times to obtain wild-type templates with concentrations of 1000 copies / μL, 100 copies / μL, 10 copies / μL, and 1 copy / μL; Method sensitivity, 10-fold gradient dilution of homozygous mutant template 2 to obtain mutant templates at concentrations of 1000 copies / μL, 100 copies / μL, 10 copies / μL, and 1 copy / μL.
[0032] 25 μL PCR reaction system contains 5 μL human genome template, 75 mmol / L Tris-HCl pH 9.0, 20 mmol / L (NH4) 2SO4 , 0.01% Tween 20, 50 mmol / L KCl, 1 U hot start Taq enzyme, 3.5 mmol / L Mg2+, 0.2 μmol / L detection probe, 0.0...
Embodiment 2
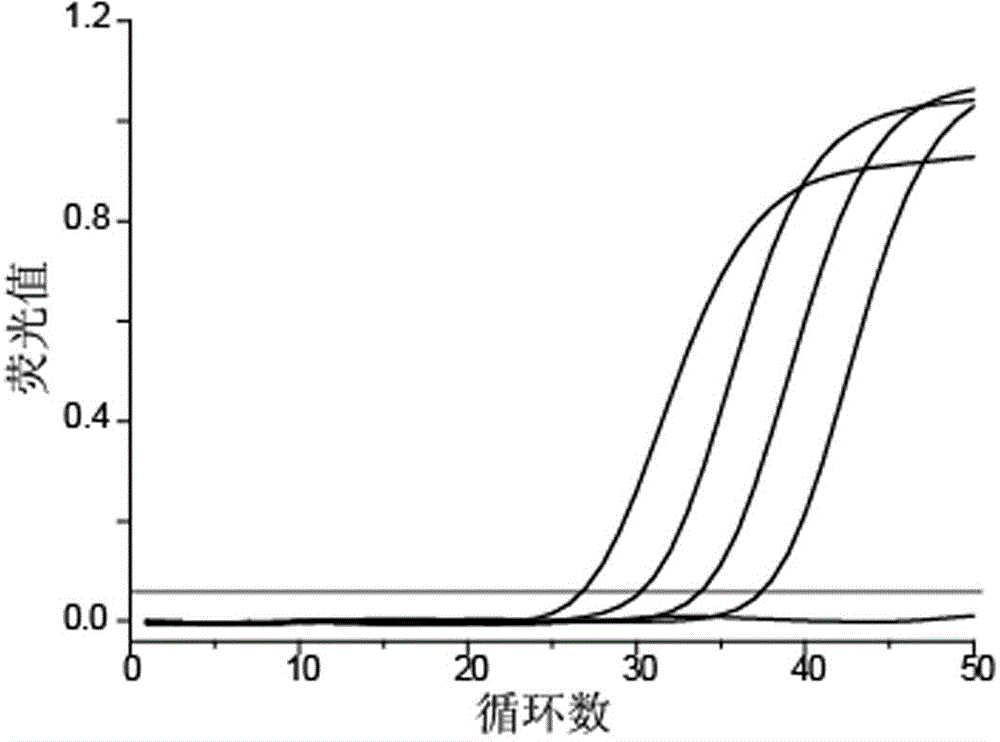
[0035] The incorporation experiment for simulating the detection of rare mutant templates is as follows: dilute a group of mutant templates with concentrations of 2000 copies / μL, 200 copies / μL, 20 copies / μL, and 2 copies / μL, and mix the mutant templates with In an equal volume of the wild-type template at a concentration of 2000 copies / μL, the ratios of mutant and wild-type templates were 1:1, 1:10, 1:100, and 1:1000, respectively.
[0036] 25 μL PCR reaction system contains 5 μL human genome template, 75 mmol / L Tris-HCl pH 9.0, 20 mmol / L (NH4) 2SO 4 , 0.01% Tween 20, 50 mmol / L KCl, 1 U hot start Taq enzyme, 3.5 mmol / L Mg2+, 0.2 μmol / L detection probe, 0.04 μmol / L upstream primer, 0.4 μmol / downstream primer. The PCR reaction program was: 3 minutes at 95°C; 50 cycles of 15 seconds at 95°C, 25 seconds at 52°C, and 20 seconds at 72°C; fluorescence signals were collected during the annealing stage at 52°C.
[0037] image 3 It shows that the template amplification curves of the ...
Embodiment 3
[0039] The 12th codon gene mutation of the K-ras gene was selected as the research object. The codon wild type is GGT, and the mutant type is CGT. According to the experimental needs, the gene sequence is synthesized as follows:
[0040] Wild type template:
[0041] TCTGAATTAGCTGTATCGTCAAGGCACTCTTGCCTACGCCACCAGCTCCAACTACCACAAGTTTATTCAGTCATTTTCAGCAGGCCTTAATAAAAATAATGAAAATGTGACTATATTAGAACATGTCACACATAAGGTTAATACACTATCAAAATACTCCACCAGTACCTTTAATA
[0042] Mutant template:
[0043] TCTGAATTAGCTGTATCGTCAAGGCACTCTTGCCTACGCCACGAGCTCCAACTACCACAAGTTTATATTCAGTCATTTTCAGCAGGCCTTATAATAAAAATAATGAAAATGTGACTATATTAGAACATGTCACACATAAGGTTAATACACTATCAAAATACTCCACCAGTACCTTTAATA
[0044] The primers and probe sequences were designed as follows:
[0045] Upstream primers: GGTGGCGTAG GTATCGTCAAGGCACTCTTGC
[0046] Downstream primer: GGAGTATTTGATAGTGTATTAACCTTATGT
[0047] Detection probe: FAM-CTCCAACTACCACAAAGTTTATATTCAGTC-BHQ
[0048] Homozygous wild-type template 3 was serially diluted 10 times to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com