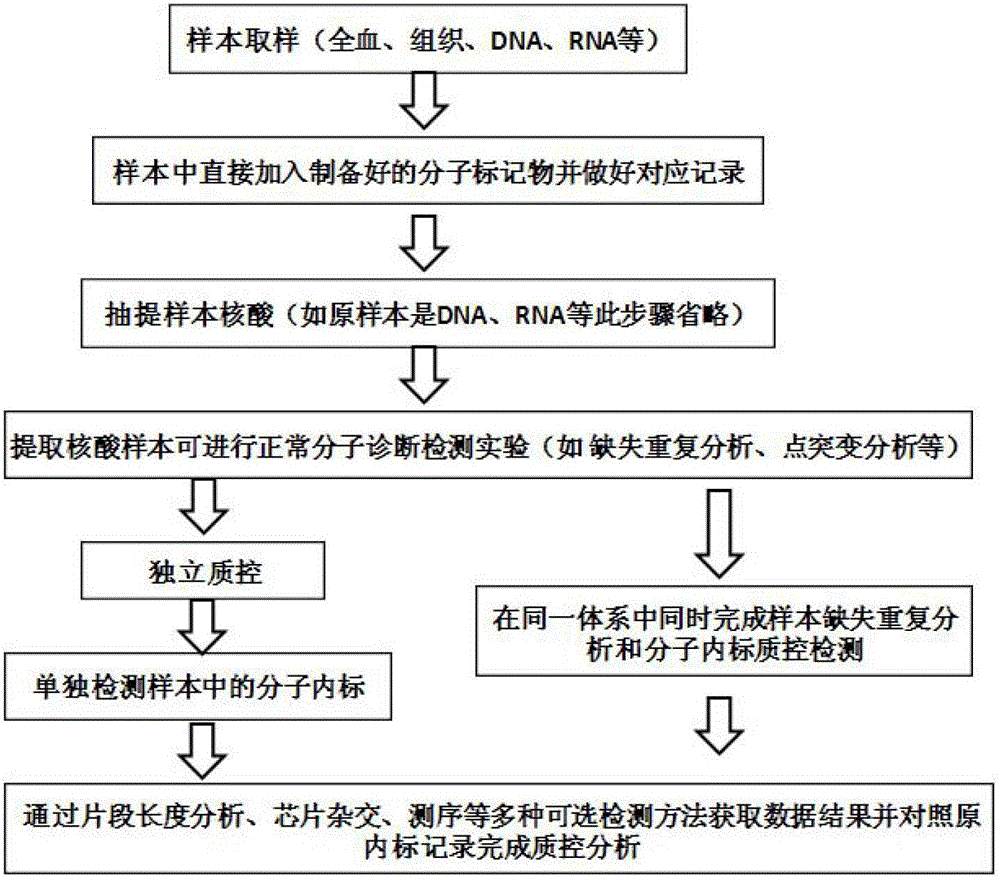
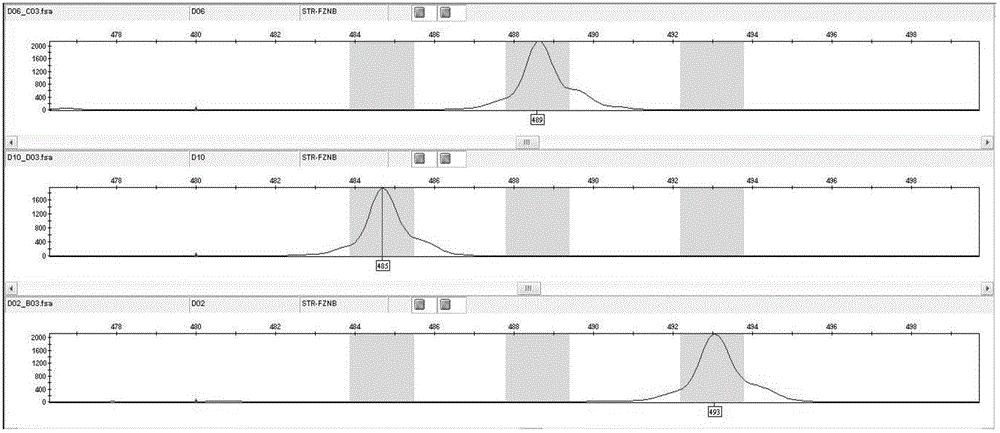
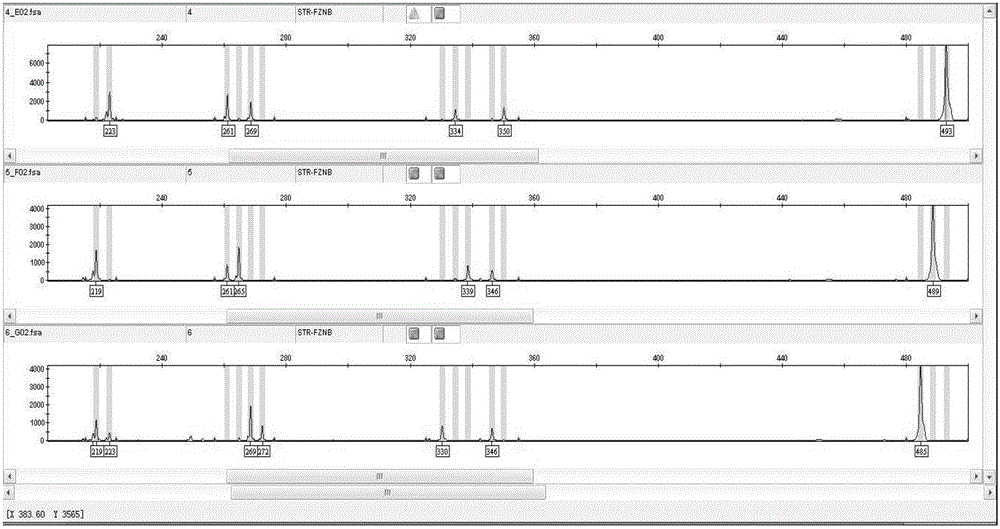
Molecular internal standard quality control method and kit for biological sample nucleic acid detection
An internal molecular standard, biological sample technology, applied in the field of biochemical detection technology and clinical molecular diagnosis, to achieve the effect of improving quality control reliability, high accuracy and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Example 1. Preparation of molecular internal standard
[0057] 1.1 Design of the original sequence
[0058] Using Random DNA Sequence online biology software (http: / / www.bioinformatics.org / sms2 / random_dna.html) to design a 1380bp DNA sequence, as shown in Seq No.1, the GC content of this sequence is between 0.4 and 0.6 Among them, after comparison of genome homology in the NCBI database, it was confirmed that there is no homologous sequence above 40 bp in a row, which means that the designed sequence does not exist in any organism.
[0059] 1.2 Site-directed mutation of Seq No.1 sequence
[0060] The method of artificially synthesizing DNA synthesizes this sequence as a template, and uses site-directed mutagenesis technology to change the bases at specific positions in the DNA sequence to achieve the effect of sequence marking, that is, use site-directed mutagenesis technology to design 6 kinds of missing bases at specific positions ( Deletion of 2bp, 4bp, 6b...
Embodiment 2
[0063] Example 2. Molecular internal standard labeling method
[0064] The sample is human whole blood as an example:
[0065] 2.1 Source or preparation of whole blood samples
[0066] The hospital can draw blood in the blood collection tube, without any follow-up processing after collection;
[0067] 2.2 Preparation of deletion-type internal standard sequences I, II, and III
[0068] 2.3 Molecular internal standard labeling method
[0069] (1) Proportionally add 0.05ng of missing molecular internal standards I, II, and III to the 200ul whole blood samples in the original tubes of the three samples, make a record of the correspondence between the samples and the internal standards, and then perform the following DNA extraction steps (using QIAamp DNA Mini Kit from QIAGEN);
[0070] (2) Add 20ul QIAGEN protease or proteinase K to the bottom of a 1.5ml centrifuge tube;
[0071] (3) Add 200ul samples (whole blood, plasma, serum, buffy coat, or 5×106 lymphoc...
Embodiment 3
[0082] Example 3. Independent internal standard quality control detection
[0083] When the internal standard in the sample needs to be detected separately, take 1ul sample and perform PCR reaction with a total reaction volume of 10 microliters (1μL 10× PCR buffer ( Qiagen company), 0.2μL 25mmol magnesium chloride, 1μL detection internal standard primer, 0.8μL 2.5mmol dNTP, 0.04 µL HotStarplus Taq enzyme ( Qiagen company), 5.96 μL wxya 2 o ). The PCR reaction conditions were set as follows: 95°C for 10 minutes; 7 cycles of Touchdown program (94°C for 20 seconds, 65°C-1 for 40 seconds and 72°C for 2 min), 28 cycles of amplification (94°C for 20 seconds, 63 ℃ 30 seconds and 72°C 2min), extended at 72°C for 2 minutes; the sequence of the missing internal standard primer is shown in Seq No.16.17:
[0084] IMMLABF: 5′-TTTTATCATGGCGCCCTCACTT-3′,
[0085] IMMLABR5'-CCTTAGCGAAGGGCGTAGGAGT-3'.
[0086] After the PCR reaction is completed, dilute the PCR amplific...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com