Agrobacterium tumefaciens mediated RNA (Ribonucleic Acid) interfering method for filamentous fungi genes
A technology of Agrobacterium tumefaciens and RNA interference, which is applied in the field of RNA interference, can solve the problems of limited application range of interference vectors, low interference efficiency, and limited application range, and achieve the effects of stable transformants, easy operation, and good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0047] 1. Preparation of competent cells: For specific experimental steps, refer to "Molecular Cloning Experiment Guide" (third edition).
[0048] 2. Transform competent cells: For specific experimental steps, refer to "Molecular Cloning Experiment Guide" (third edition).
[0049] 3. Enzyme digestion method: for parameters such as the amount of restriction endonuclease used, the choice of buffer, and the enzyme digestion system, refer to the instruction manual attached to Dalian Takara Company;
[0050] 4. Plasmid extraction method: refer to the instructions attached to the plasmid extraction kit (Tiangen Biochemical Technology Co., Ltd.)
[0051] 5. The method of purifying PCR products: refer to the instructions attached to the DNA purification and recovery kit (Tiangen Biochemical Technology Co., Ltd.)
[0052] 6. Ligation of PCR products and vector fragments: For the usage of DNA ligase and Taq enzyme, refer to the instructions attached to Dalian Takara Company.
[0053]...
Embodiment 1
[0065] The construction of embodiment 1 PCB309
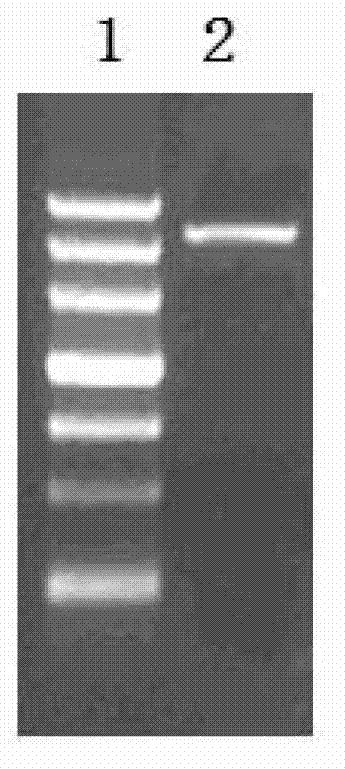
[0066] 1. Construction of pCB298: Using pBI121 as a template, use primers PCB298-F and PCB298-R to amplify the RK2 replication region and nptⅡ, with a length of 4566bp. The reaction conditions were pre-denaturation at 94°C for 1 min; denaturation at 94°C for 30 sec, annealing at 55°C for 45 sec, extension at 72°C for 4 min, 30 cycles; 10 min at 72°C. The PCR fragment was purified and recovered, digested with XbaI, purified and recovered after digestion. The purified fragments after enzyme digestion were ligated, and the single fragments were ligated into circular plasmids. The ligation product was transformed into Escherichia coli DH5α, a single colony was picked, the plasmid was extracted, and verified by XbaI single enzyme digestion.
[0067] Recombinant vector pCB289 enzyme digestion identification picture figure 1 : Among them, 1 is marker: DL4500; 2 is the 4566bp PCB289 digestion product; the digestion product is consist...
Embodiment 2
[0116] Example 2. Construction of RNA interference vector pCB309-pfgrt
[0117] 1. Construction of pUC-pgt
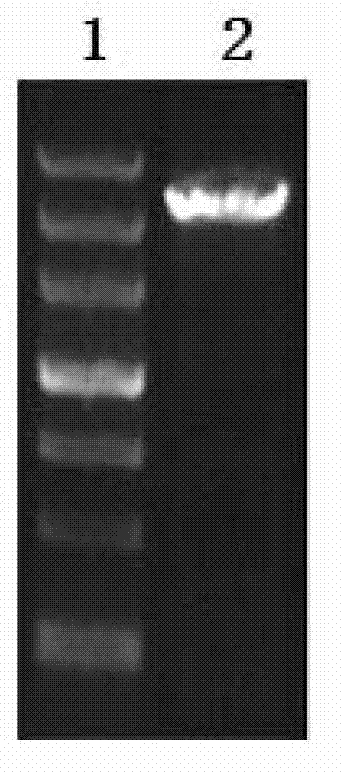
[0118] Using the pSilent-1 plasmid as a template, primers PUT-F and PUT-R were used to amplify the sequences of the Ptrpc promoter, Gus spacer sequence and Ttrpc terminator, and the fragment length was about 2.2 kb. The reaction conditions were pre-denaturation at 94°C for 1 min; denaturation at 94°C for 30 sec, annealing at 55°C for 45 sec, extension at 72°C for 3 min, 30 cycles; 10 min at 72°C. The purified PCR fragment was connected to the Simple T vector, and connected into a circular plasmid to obtain pUC-pgt. The ligation product was transformed into Escherichia coli DH5α, a single colony was picked, the plasmid was extracted, PCR was verified with primers PUT-F and PUT-R, and the positive clone was named as the recombinant vector pUC-pgt.
[0119] PCR identification map of recombinant vector pUC-pgt; Figure 5 : Among them, 1 is marker: DL4500; 2 is the PUC-pg...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com