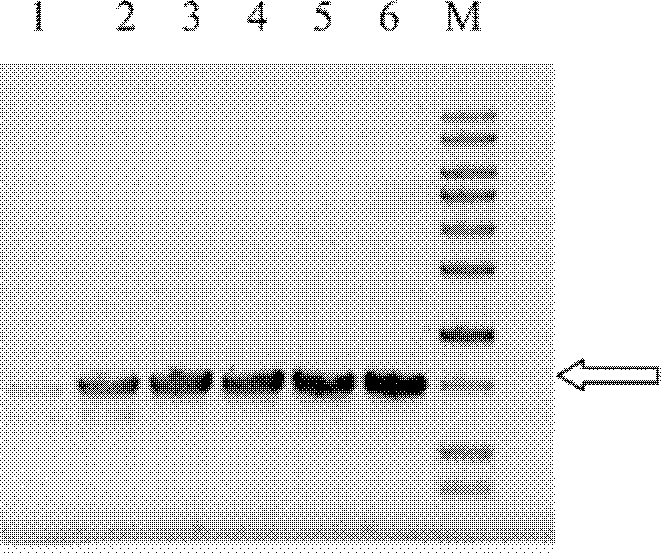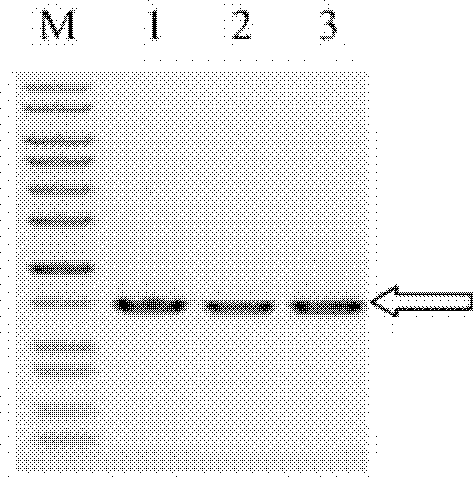Chondrosulphatase B fusion protein, and coding gene and construction method thereof
A technology of chondroitin sulfate enzyme and chondroitin sulfate, which is applied in the field of genetic engineering and fermentation engineering, can solve the problems of inability to increase protein solubility, weak affinity with affinity carrier, and difficulty in improving the purification effect of protein soluble ratio, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0104] Embodiment 1, expression of chondroitin sulfate B fusion protein MBP-ChSase B
[0105] 1. Cloning of Flavobacterium heparinus chondroitinase B coding sequence with signal peptide removed
[0106] The specific process of the construction of the expression vector pMAL-ChSase B is as follows:
[0107] 1. Design and synthesis of primers
[0108] The DNA sequence of Flavobacterium heparinus chondroitinase B was queried through Genbank (Tkalec, A.L., Fink, D., Blain, F., Zhang-Sun, G., Laliberte, M., Bennett, D.C., Gu, K. , Zimmermann, J.J. and Su, H. Isolation and expression in Escherichia coli of cslA and cslB, genes coding for the chondroitin sulfate-degrading enzymes chondroitinase AC and chondroitinase B, respectively, from Flavobacterium heparinum.Appl.Environ.Microbiol6.2000, (1), 29-35), and then design primers according to the DNA sequence of Flavobacterium heparinus chondroitinase B that encodes the signal peptide base, and introduce the recognition sites of res...
Embodiment 2
[0127] Embodiment 2, by optimizing the concentration of the inducer IPTG and the composition of the medium to improve the softness of sulfate Expression and activity of osteosinase B fusion protein MBP-ChSase B
[0128] The optimal expression host E.coli TB1 / pMAL-ChSase B in Example 1 was selected, and by optimizing the concentration of the inducer IPTG, the expression level of the chondroitinase B fusion protein MBP-ChSase B was greatly improved, and the Enzyme activity per unit of fermentation broth.
[0129] Bacterial culture, crushing, protein quantity determination and enzyme activity determination are the same as Step 3 of Example 1. The obtained chondroitin sulfate B fusion protein can be directly used to measure the activity of chondroitin sulfate B, and the part of maltose binding protein is not excised.
[0130] Figure 6 The results of optimization of the inducer IPTG concentration are shown. It can be found that the optimal IPTG concentration for inducing th...
Embodiment 3
[0132] Example 3, Purification of Chondroitinase B Fusion Protein MBP-ChSase B by Amylose Column
[0133] The fusion partner (fusion partner) maltose binding protein MBP utilized in the present invention can achieve one-step separation with amylose affinity adsorption. The specific steps of affinity separation are as follows: 100 mL of bacterium whose final concentration was 0.24 mM IPTG induced expression for 23 hours was centrifuged at 10,000 rpm for 10 minutes; at the same time, a bacterium with no induced expression was set as a control. Then proceed with the following two options:
[0134]Option 1: Wash twice with Column buffer (20mM Tris-HCl, 200mM NaCl, pH7.5), resuspend in 5mL Column buffer, and perform sonication (300W output power, 3 seconds each time and intermittent 99 times in 3 seconds).
[0135] Option 2: Osmotic pressure shock. Resuspend the bacteria in 100 mL osmotic shock buffer I (20-40% sucrose, 30 mM Tris-HCl, 1 mM EDTA) for 15 minutes, and stir. Cen...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com