One-step loop-mediated reverse transcription isothermal amplification detection method for soybean mosaic virus
A soybean mosaic virus and isothermal amplification technology, applied in the field of warm amplification detection, can solve the problems of not being used for rapid detection, and achieve the effects of saving redundant time, simplifying the operation process, and speeding up the inspection speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 Soybean mosaic virus RT-LAMP primer set design and screening:
[0038] By aligning the SMV coat protein (CP) sequences of different strains in GenBank, find the SMV CP conserved region sequence shown in SEQ ID NO.5 (GenBank: HM590055.1) as the template reference sequence, using online LAMP Primer design software primerExplorer V4 ( http: / / primerexplorerjp / elamp4.0.0 / index.html) Four sets of primer sets were designed, and the primer sets are as follows:
[0039] Primer set I:
[0040] F3-I: GCCATTAGCATCTGGAGAT (SEQ ID NO. 6)
[0041] B3-I: CTTGCTTGAGTACAAACCTAA (SEQ ID NO. 7)
[0042] FIP-I: ACGATGAGCAGATGGGTGTGTACCATTGTCAATGCACCATA (SEQ ID NO. 8)
[0043] BIP-I: TCTTTAACTGCATTGTACCACGCGGTTGATTTATTCAACACTCGA (SEQ ID NO. 9)
[0044] Primer Set II:
[0045] F3: AGATGTAAATGTTGGATC (SEQ ID NO. 1)
[0046] B3: TCATCATCAAGCTCATATT (SEQ ID NO. 2)
[0047] FIP: ATCTTCCCTTCAACCATTGGAAGAAGGTGGTTCCGCGTTTGCAGAAG (SEQ ID NO. 3)
[0048] BIP: CCTAATCAGGTTGATTTATTCA...
Embodiment 2
[0065] Example 2 Optimization experiment of soybean mosaic virus RT-LAMP reaction system
[0066] (1) To optimize the concentration of Mg ions, add 25 mM MgSO in sequence according to the reaction system 4 : 0.8, 1.2, 1.6, 2.0, 2.4, 2.8ul.
[0067] (2) To optimize the betaine concentration, add 5M betaine in sequence according to the reaction system: 0.8, 1.2, 1.6, 2.0, 2.4, 2.8ul.
[0068] (3) To optimize the dNTP concentration, add 10 mM dNTP according to the reaction system: 0.4, 0.6, 0.8, 1.0, 1.2, 1.4ul.
[0069] (4) Optimize the concentration of inner primers (FIP, BIP), and add them in sequence according to the reaction system (FIP, BIP): 0.2, 0.3, 0.4, 0.5, 0.6, 0.7. At this time, the outer primers were all used in 0.2ul.
[0070] The original reaction system (see Example 1) was used for the control group, the control without any RNA was used as the blank control, the plasmid was used as the positive control, and the total RNA extracted from soybean samples with SMV...
Embodiment 3
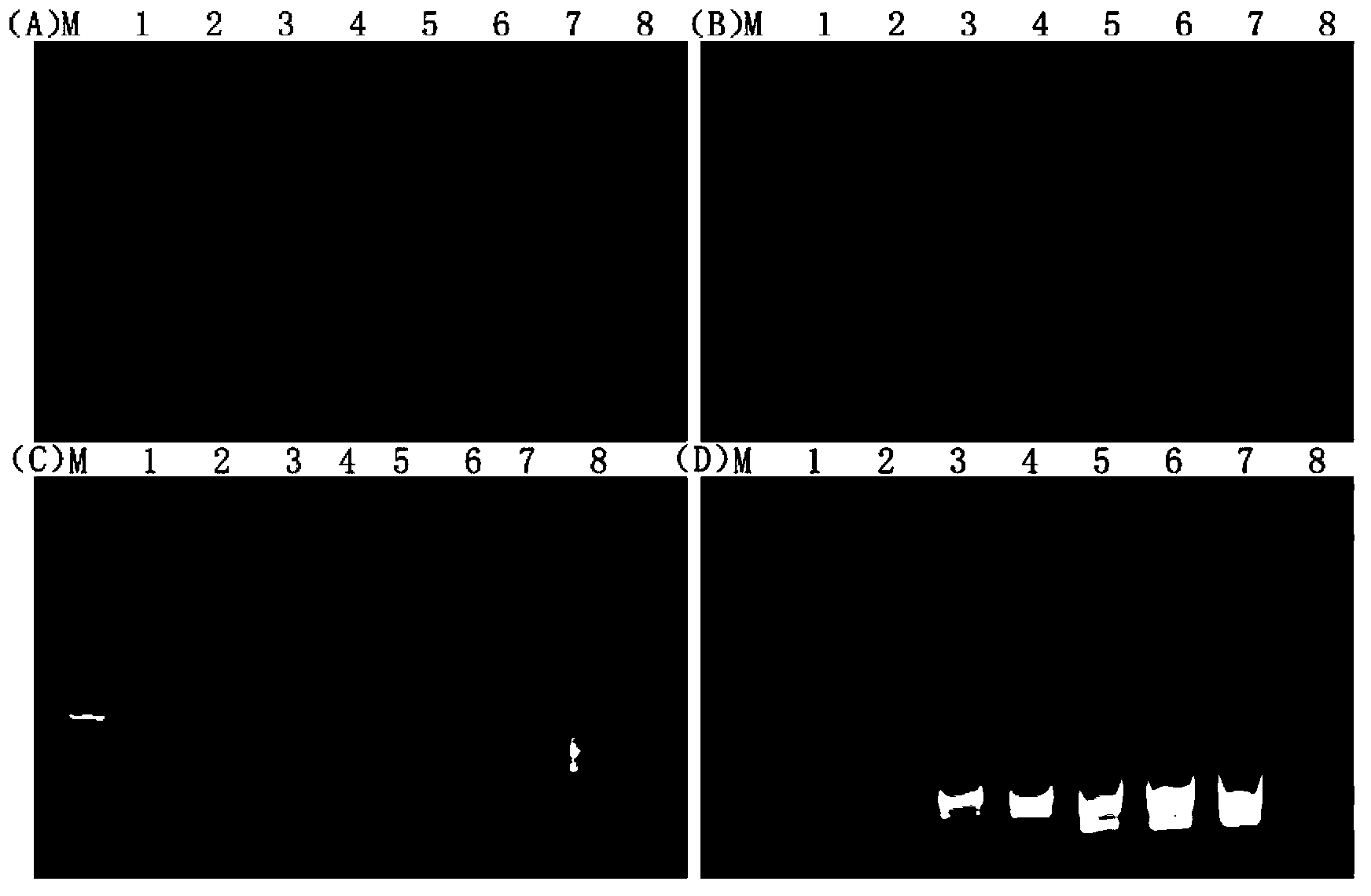
[0075] Embodiment 3 RT-LAMP and RT-PCR sensitivity comparison
[0076] In order to determine the sensitivity of the two detection methods RT-LAMP and RT-PCR, the concentration of the extracted total RNA was measured with a spectrophotometer, and RNAse-Free ddH was used to measure the concentration of the total RNA. 2 O dilution and stored at -70°C as template. Perform 10 steps on 100ng of RNA template in sequence -1 It was diluted to 0.001ng, 1ul RNA was used as template, and primer set II was used to carry out the reaction according to the system in Example 2. Take 2ul of the amplified product as sample and run it on a 1% agarose gel. At the same time, the same template of RT-LAMP was used, B3 was the reverse transcription primer, and the first strand of cDNA was synthesized by reverse transcription according to the instructions of AMV reverse transcriptase. The cDNA synthesized by RT was used as a template, and F3 and B3 were used as primers for PCR amplification, and a c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com