Bacterial strain for producing farnesene and application of bacterial strain
A technology of farnesene and strains, applied in the field of synthetic biology, can solve problems such as failure to implement industrial production and unsatisfactory protein expression, and achieve good industrial application prospects, increase production, and experimental rationality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
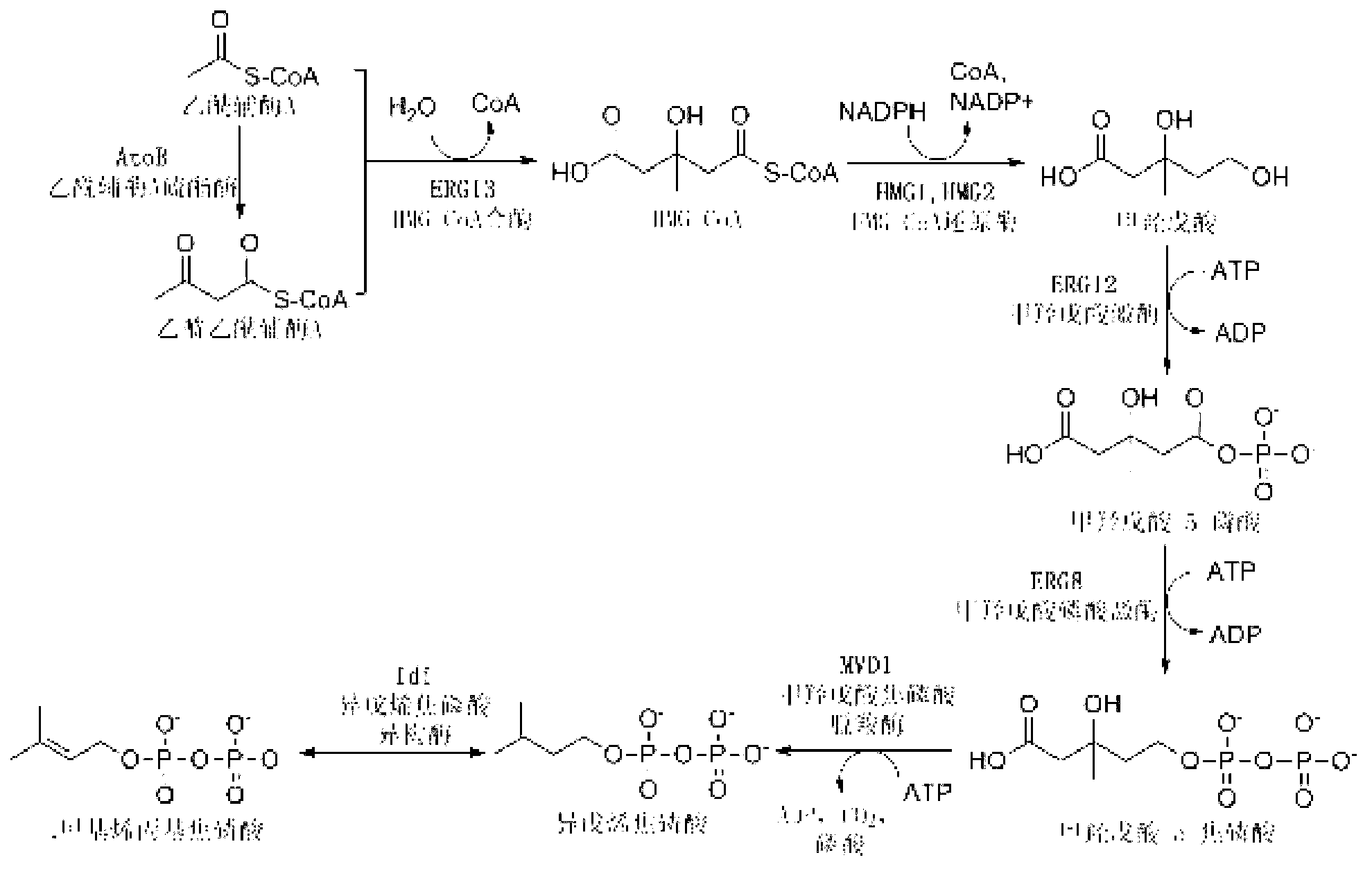
[0039] Example 1 In Vitro Reconstruction System Optimization of Mevalonate Pathway
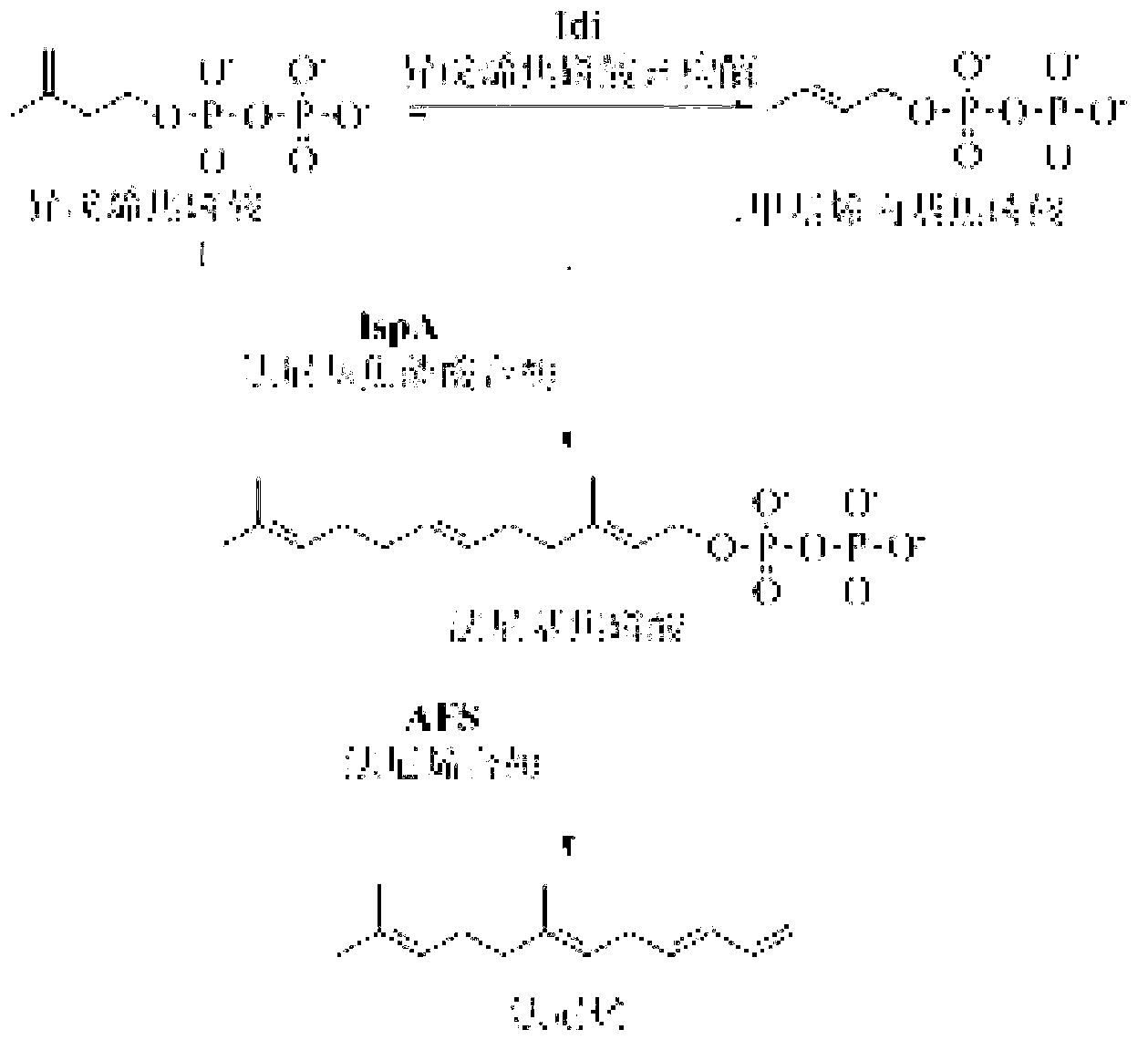
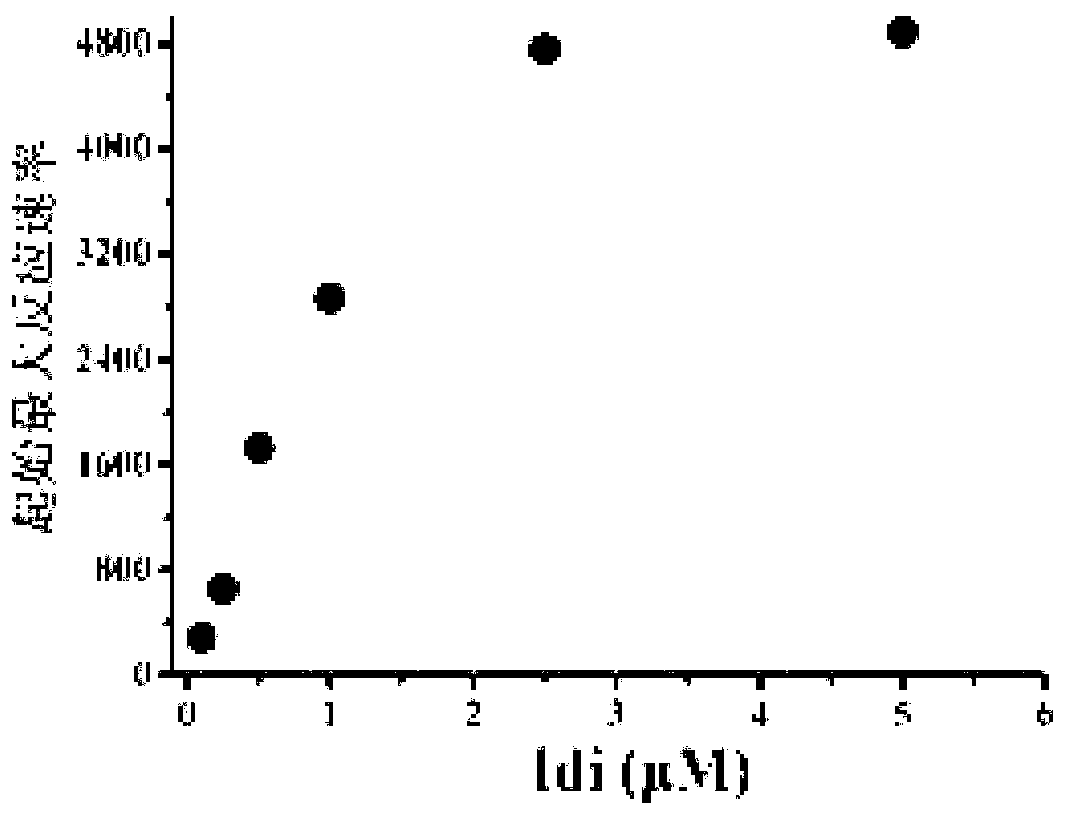
[0040] The experimental method of the in vitro reconstitution system is the same as that established by Yu et al. in 2011 (Yu, X., et al., 2011. .108,18643-18648.), through the study of each protein in the mevalonate pathway, it was shown that increasing ERG13, ERG8, IspA and Idi can promote the production of terpenoids, especially the effect of increasing Idi is more obvious ( image 3 ), the optimized ratio of each protein in the synthesis of farnesene in the MVA pathway obtained from the in vitro reconstruction experiment is AtoB:ERG13:tHMG1:ERG12:ERG8:MVD1:Idi:IspA:AFS=1:10:2:5:5:2:5 : 2: 2, compared with the equimolar amounts of each protein, the reaction rate is significantly improved ( Figure 4 ).
Embodiment 2
[0041] Embodiment 2 Mevalonate pathway plasmid construction
[0042] Escherichia coli BL21(DE3) genomic DNA and Saccharomyces cerevisiae INVSC1 genomic DNA were purified with Qiagen's Blood and Cell Culture DNA Mini Kit.
[0043] Plasmid pMH1 contains the first three genes of the mevalonate pathway: atoB gene (acetoacetyl-CoA thioesterase) from Escherichia coli BL21 (DE3), erg13 (HMG-CoA synthase, 3- hydroxymethyl-glutaryl-CoA synthase) and thmg1 (HMG-CoA reductase, 3-hydroxymethyl-glutaryl-CoA reductase, deletes the transmembrane region of HMG1). Plasmid pFZ81 contains the last four genes of the mevalonate pathway: erg12 (mevalonate kinase), erg8 (mevalonate-5-phosphate kinase) and mvd1 (mevalonate-5-phosphate kinase) from Saccharomyces cerevisiae INVSC1 pyrophosphate kinase), derived from the idi (isopentenyl pyrophosphate isomerase) gene of Escherichia coli BL21 (DE3). All genes were amplified by PCR, and the primers used are listed in Table 1.
[0044] The specific cons...
Embodiment 3
[0050] Example 3 Farnesene produces metabolic pathway plasmid construction
[0051] The afs gene from Malus x domestica was codon-optimized by Nanjing KingScript Biotechnology Co., Ltd. for gene synthesis (sequence shown in SEQ ID NO.1), and then inserted into the plasmid through NdeI and XhoI restriction sites In pET21a(+), the plasmid pFZ35 ( Figure 7 ).
[0052] IspA gene was amplified from E. coli genomic DNA with primers IspA-BamHI and IspA-EcoRI-SpeI, digested with BamHI and EcoRI and inserted into plasmid pET28a(+) to obtain pFZ32. Then insert the XbaI-XhoI fragment containing ispA derived from pFZ32 into the SpeI-XhoI fragment of pFZ35 to obtain plasmid pFZ38, the plasmid schematic diagram is as follows Figure 8 As shown, the sequence (excluding the backbone vector sequence) is shown in SEQ ID NO.4.
[0053] The Idi gene was amplified by PCR using BL21(DE3) genomic DNA as a template (see Table 1 for primers Idi-NdeI and Idi-XhoI), and then digested with NdeI and X...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com