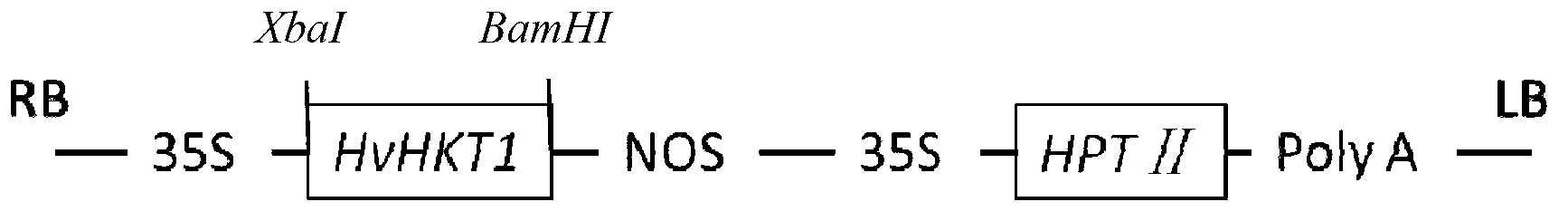Application of barley HvHKT1gene
A gene and barley technology, applied in the field of plant functional genomics, can solve the problems of normal growth and development of transgenic plants, improvement of salt tolerance, little knowledge of environmental stress tolerance genes, and far-flung requirements, so as to reduce sodium ions. The accumulation, obvious practicability and application prospects, the effect of increasing production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Embodiment 1, the cloning of HvHKT1 coding gene
[0023] Wild barley X16 seeds treated with 3% (v / v) H 2 o 2After surface disinfection for 20 minutes, rinse with distilled water three times, sow on moist double-layer filter paper, and germinate in a growth chamber at 20°C for 10 days (14 hours of light / 10 hours of darkness). Cut the young leaves, put them in a mortar and add liquid nitrogen to grind them, extract the total RNA of the tissue with reference to the Tiangen Plant RNA Extraction Kit (DP432), and reverse transcribe to obtain the first-strand cDNA (Takara Reverse Transcription Kit RR037Q), using this cDNA as a template, use
[0024] Primer KTI-F1 (5'-attTCTAGAatgcctgaactcgaaagccc-3')
[0025] PCR amplification was performed with primer KTI-R1 (5'-attGGATCCctaatttgcagccacctctgg-3'), and the kit was from Quanshijin Company (AP131).
[0026] The 50μL reaction system is:
[0027] Transgen HiFi system
[0028] The reaction program was: 95°C for 5 min...
Embodiment 2
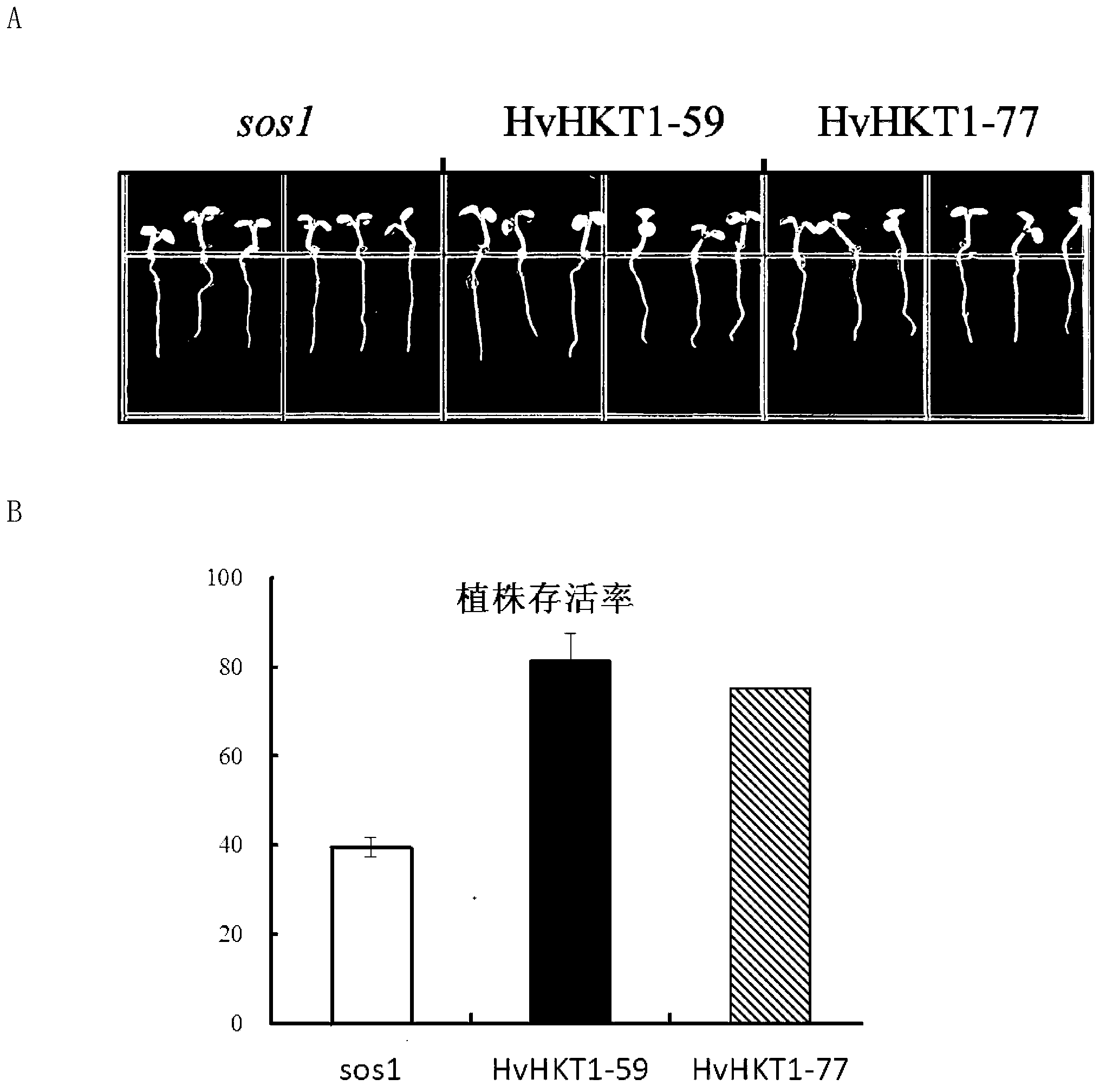
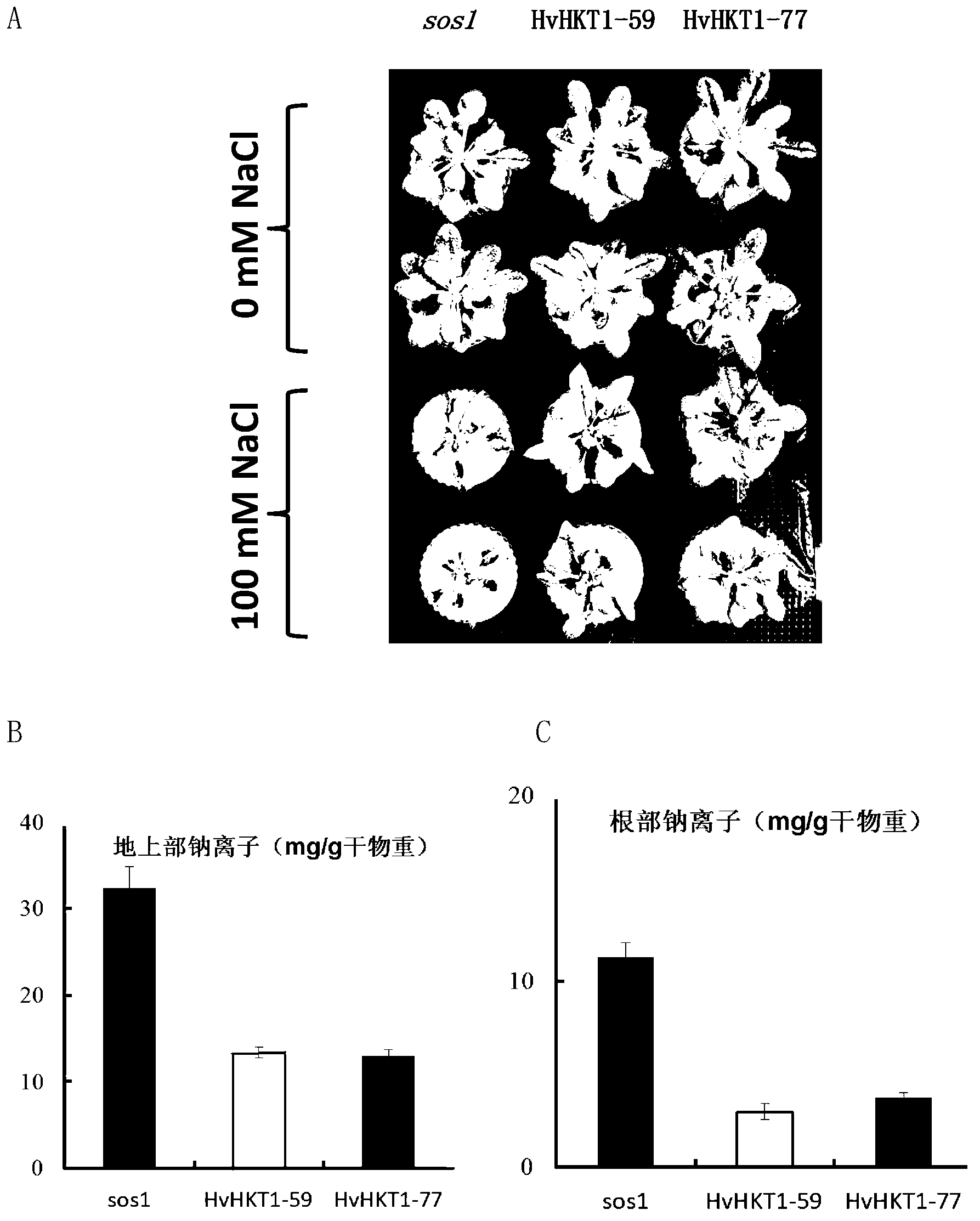
[0029] Example 2. Application of HvHKT1 gene to enhance plant salt tolerance
[0030] 1. Construction of overexpression vector in Arabidopsis
[0031] The coding sequence of HvHKT1 was cloned into pCambia 1301 vector and placed under the drive of 35S promoter. The specific method is as follows: the fragment amplified in Example 1 and the pCambia 1301 plasmid vector were double digested with BamHI and XbaI, and the gene fragment was inserted between the BamHI and XbaI enzyme recognition sites of the vector using T4 DNA ligase to obtain p1301-HvHKT1 recombinant vector. After the recombinant vector was sequenced and identified by multi-enzyme digestion, it was shown that the HvHKT1 gene was correctly connected to the pCambia 1301 vector.
[0032] 2. Transformation of Arabidopsis thaliana with p1301-HvHKT1
[0033] The recombinant vector p1301-HvHKT1 was transformed into Agrobacterium GV3101 competent by the freeze-thaw method, and a positive single colony was picked on the LB ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com