Medicolegal identification method of species
A technology of species identification and identification method, which is applied in the field of forensic species identification, can solve the problems of misidentification, lower sensitivity, and cumbersome detection process, etc., and achieve the effect of simple identification process, guaranteed accuracy, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
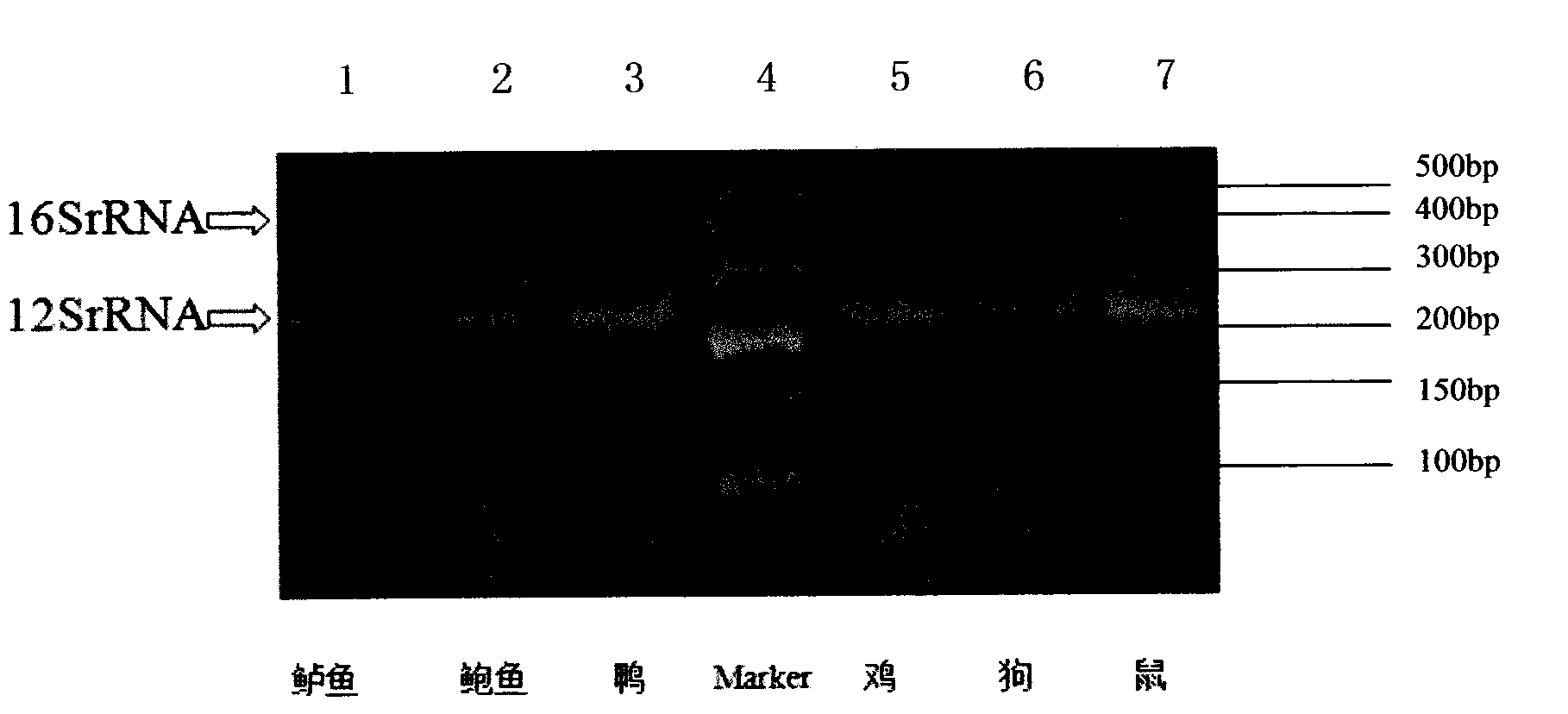
[0045] Example 1: Identification of human and animal samples:
[0046] Extract mtDNA (mitochondrial DNA) from human and rhesus monkeys, chimpanzees, macaques, sheep, pigs, cows and rabbits and other 7 kinds of animal tissues or blood, and divide them into 8 experimental groups from 1 to 8 in turn. The amplification system of PCR amplification is 1 μl of template, 3.2 μl of primers, 2.5 μl of 10× buffer, 2 μl of dNTP, 0.25 μl of Taq enzyme and 16.05 μl of deionized water. The whole amplification system is 25 μl; the primers of each amplification system All are 12SF1 / 12SR, 12SF2 / 12SR, COX1F / COX1R and 16SF / 16SR mixtures, the 3.2 μl primers contain 7 primers, and the primer concentration is 10 μM, of which 12SF1 is 0.3 μl, 12SF2 is 0.3 μl, and 12SR is 0.6 μl, 0.5 μl for 16SF, 0.5 μl for 16SR, 0.5 μl for COX1F, and 0.5ul for COX1R; primer sequences were synthesized by Treasure Bioengineering (Dalian) Co., Ltd., OPC purity; Taq enzyme was EasyTaq DNA polymerase from Transgen.
[...
Embodiment 2
[0059] Example 2: The sensitivity experiment of identification method of the present invention:
[0060] 1. Determination of DNA amount: after extracting mtDNA according to the method for extracting mtDNA in Example 1, quantify with spectrophotometry and measure OD 260 , according to the calculation method of double-stranded DNA1OD corresponding to 50ug / ml, quantify, dilute mtDNA with water to 2.5pg, 5pg, 10pg, 20pg, 50pg, 100pg, lng,
[0061] 2. According to the method in Example 1, carry out PCR amplification to the mtDNA of 7 kinds of concentrations in step (1);
[0062] 3. Carry out agarose gel electrophoresis according to the method in Example 1.
[0063] The result is as image 3 As shown, the amount of DNA template in wells 1, 2, 3, 4, 5, 6, and 7 from left to right is 2.5pg, 5pg, 10pg, 20pg, 50pg, 100pg, and 1ng.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com