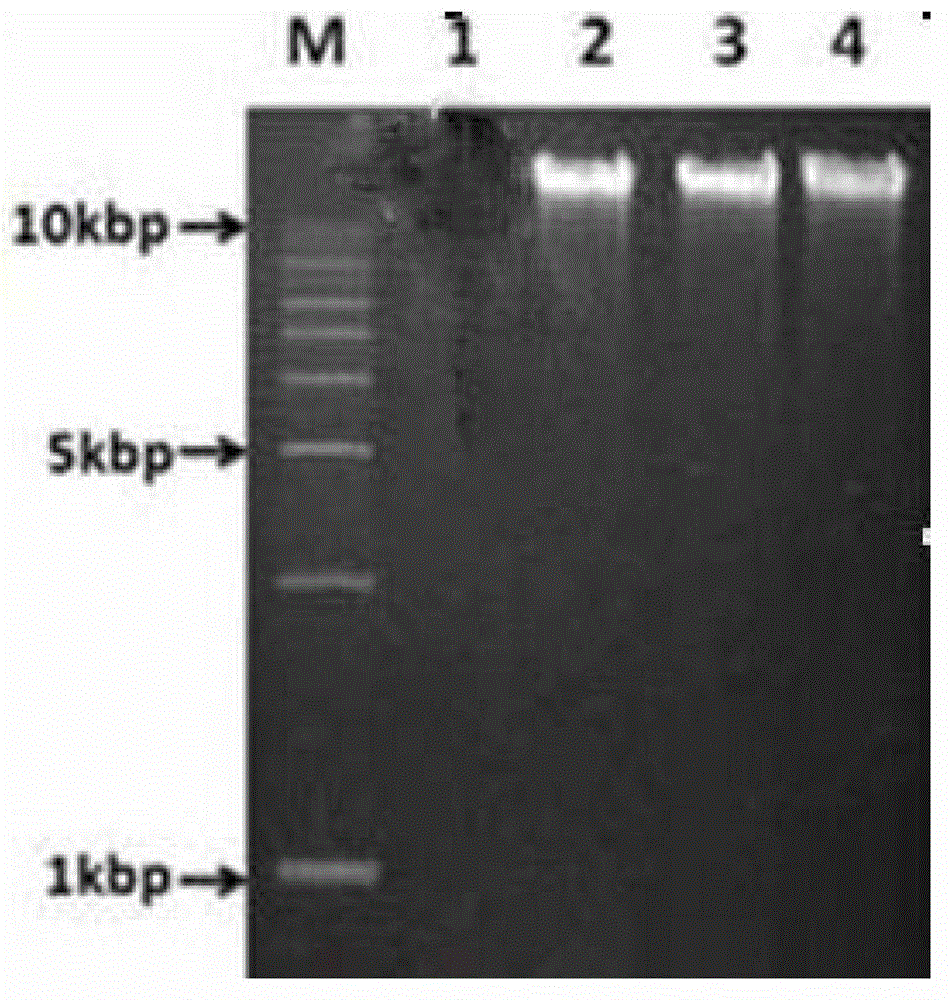
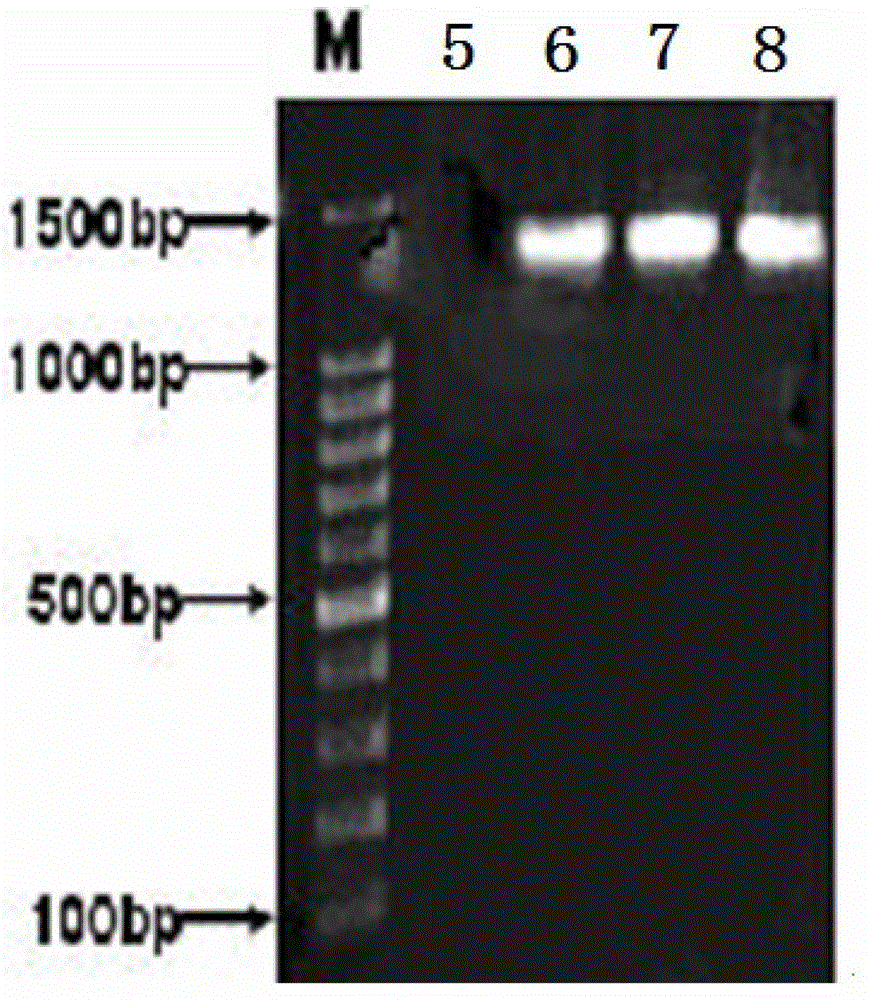
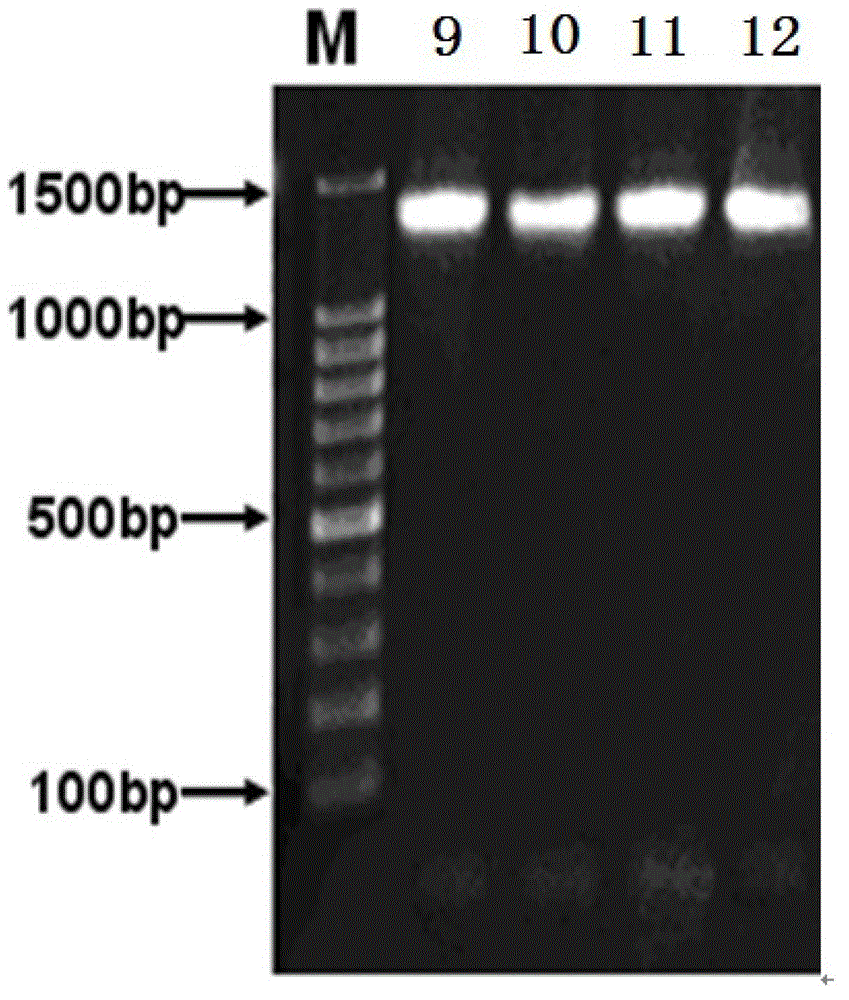
Method for efficiently extracting underground water microbial DNA for PCR amplification
A technology for microorganisms and groundwater, applied in the direction of DNA preparation, recombinant DNA technology, etc., can solve the problems of small number of microorganisms in groundwater and high extraction difficulty, and achieve the effect of simple method, low cost and low operation cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] The technical solution adopted by the present invention is a method for efficiently extracting groundwater microbial DNA for PCR amplification. The method includes the steps of collecting groundwater microorganisms, cracking groundwater microbial cells, purifying cell lysates, and extracting and purifying microbial genome DNA. The concrete steps of the inventive method are:
[0042] The first step is to collect groundwater microorganisms: take groundwater samples and filter them with sterile microporous membranes to collect microorganisms, and then use a buffer containing cetyltrimethylammonium bromide to elute the microorganisms on the sterile microporous membranes Microbial cell liquid is obtained; the buffer contains cetyltrimethylammonium bromide, which can achieve better elution effect.
[0043] The second step, cracking groundwater microbial cells: add lysozyme to the microbial cell liquid for lysozyme enzymolysis, transfer the lysate after lysozyme enzymolysis in...
Embodiment 2
[0047] On the basis of embodiment 1, the present invention is in order to improve the extraction effect of genomic DNA, preferred embodiment also has, and the operation process of cracking groundwater microbial cell described in the second step is:
[0048] ①Lysozyme enzymatic hydrolysis: add lysozyme solution to the microbial cell liquid until the concentration of lysozyme in the microbial cell liquid is 10mg / mL, put in a water bath at 35°C for 1.5h, and then transfer to a new centrifuge tube;
[0049] ②Protease hydrolysis: add proteinase K to the microbial cell fluid after lysozyme hydrolysis, until the concentration of proteinase K in the microbial cell fluid is 0.2mg / mL, then add 100 microliters of dodecyl sulfate with a mass concentration of 20% Sodium, 40 microliters of 5mol / L NaCl solution and 20 microliters of 2mol / L NaCl 3 PO 4 The solution was placed in a water bath at 53° C. for 2 hours, centrifuged at a speed of 11,000 r / min for 5 minutes, and the supernatant was ...
Embodiment 3
[0052] On the basis of Example 2, the present invention obtains high-purity genomic DNA in order to purify the cell lysate, and a preferred embodiment also has the operation process of purifying the cell lysate described in the third step as follows:
[0053] ① Add solution III which is 0.2 times the volume of the cell lysis supernatant to the cell lysis supernatant and mix evenly. After 12 minutes in ice bath, centrifuge at 12,000 r / min for 13 minutes at 4°C to remove the precipitate and obtain No. 1 supernatant;
[0054] ② Add pre-cooled absolute ethanol twice the volume of the No. 1 supernatant to the No. 1 supernatant, precipitate at -20°C for 60 minutes, then centrifuge at 12,000 r / min at 4°C for 14 minutes, and remove the supernatant After obtaining No. 1 precipitation;
[0055] ③ Add 500 microliters of extraction buffer to the No. 1 precipitate to dissolve the No. 1 precipitate to obtain a crude DNA extraction solution. Using absolute ethanol to precipitate DNA and RNA...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pore size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com