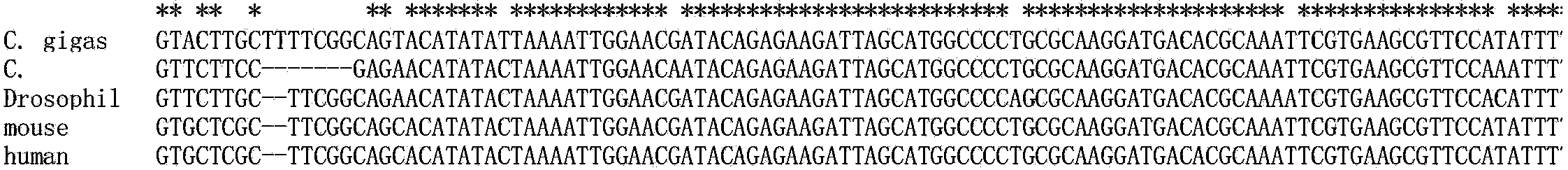Shellfish sn RNA U6 gene, primer, and applications thereof
A gene and shellfish technology, which is applied in the field of the complete sequence of shellfish snRNA U6 gene and its primers, and the expression analysis of shellfish microRNA, can solve the problems of designing primers and no U6 gene, etc., and achieves low cost, simple operation, and high sensitivity high effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] The putative Oyster U6 gene sequence was obtained from the Oyster oyster genome based on homologous sequence searches. Primers were designed in its conserved region to obtain the partial sequence of the U6 gene. According to the obtained partial sequence, the full length of the U6 gene was obtained by using RACE technology.
[0033] Sequence Listing (1) Information of SEQ ID NO 1
[0034] sequence feature
[0035] Length: 109nt
[0036] Type: nucleic acid
[0037] Chain type: double chain
[0038] Topology: Linear
[0039] Molecule type: DNA
[0040] Feature name: snRNA (1-109)
[0041] Source: Long Oyster
[0042] Sequence description:
[0043] GTACTTGCTTTTCGGCAGTACATATATTAAAATTGGAACGATACAGAGAAGATTAGCATGGCCCCTGCGCAAGGATGACACGCAAATTCGTGAAGCGTTCCATATTTTT
[0044] Based on homology alignments with other species (see figure 1 ), and found that the gene sequence was highly conserved among different species, thus confirming it as U6 gene.
Embodiment 2
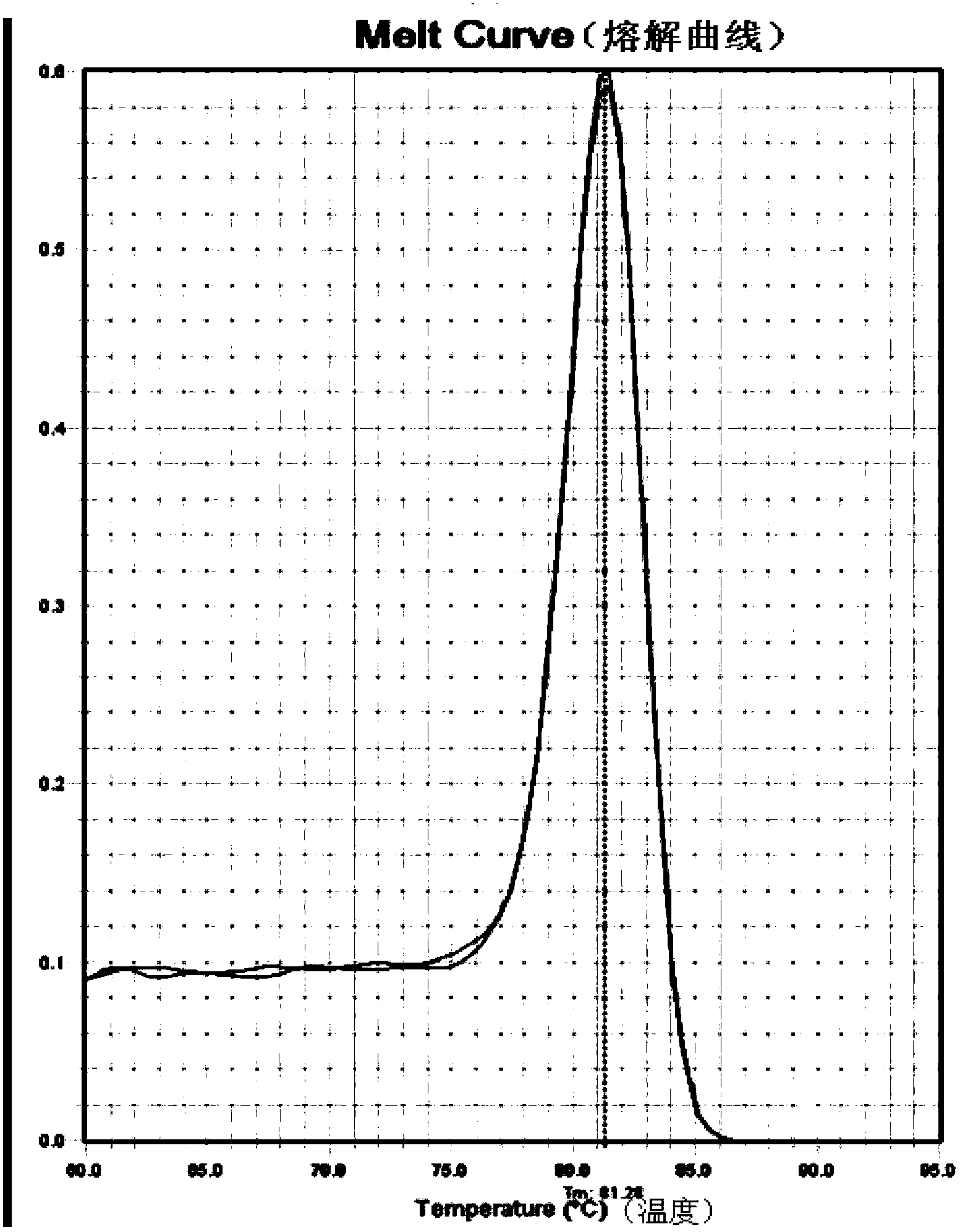
[0046] A pair of primers were designed according to the U6 gene sequence. The pair of primers has high specificity, the amplification efficiency is 1.0016, and the applicable annealing temperature range is wide (54-68°C).
[0047] Design a pair of fluorescent quantitative primers in the Primer Premier 5 software based on the full length of the U6 gene:
[0048] Forward primer: 5'-CGGCAGTACATATATTAAAATTGGAACGA-3'
[0049] Reverse primer: 5'-TGGAACGCTTCACGAATTTGCGTGTCATC-3'
[0050]This primer amplifies a 90bp PCR product:
[0051] CGGCAGTACATATATTAAAATTGGAACGATACAGAGAAGATTAGCATGGCCCCTGCGCAAGGATGACACGCAAATTCGTGAAGCGTTCCA
[0052] The specific embodiment of this pair of primer PCR amplification is as follows:
[0053] 1. Experimental materials: Oyster larvae and adult tissue gills were taken from Qingdao, Shandong. Larval samples were from F2 progeny, and F1 was 51 female oysters and 1 male oyster from the G3 line. The G3 family was constructed from a pair of male and femal...
Embodiment 3
[0080] U6 was used as an internal reference, and the method based on SYBR Green dye fluorescence quantitative PCR was used. See the literature: [Shi R, Chiang VL (2005) Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 2550 39: 519-525.] for long Oyster miRNA relative quantification.
[0081] The miRNA sequence (see Table 2) and sequencing expression profile (see Table 3) of Oyster oyster are from the data of our laboratory, by the method of deep sequencing, see the literature [Huang J, Hao P, Chen H, Hu W, Yan Q, et a1. (2009) Genome-wide identification of Schistosoma japonicum microRNAs using a deep-sequencing approach. P1os One 4: e8206,] obtained:
[0082] The experimental materials and RNA extraction method are the same as in Example 2. Small RNA sequencing was performed on the I1lumina sequencing platform of Shenzhen BGI Institute. Firstly, 18-30 nt RNAs were recovered from the sample RNA by gel cutting, and the 5’ and 3’ sequencing adapt...
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com