Animal chlamydia TaqMan-MGB probe multiplex real-time fluorescent quantitative PCR (polymerase chain reaction) detection primers, kit and method
A technology for real-time fluorescence quantification and animal chlamydia, which is applied in biochemical equipment and methods, microbial measurement/inspection, fluorescence/phosphorescence, etc., can solve the problems of detection kits and detection methods for three types of chlamydia, and achieve identification Simple, highly specific effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Embodiment 1, design and screening of primers
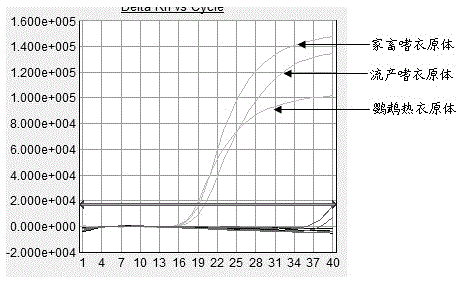
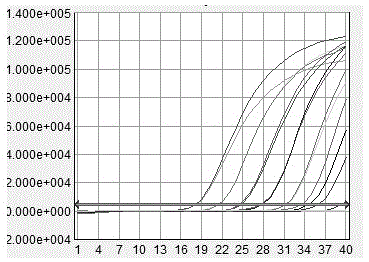
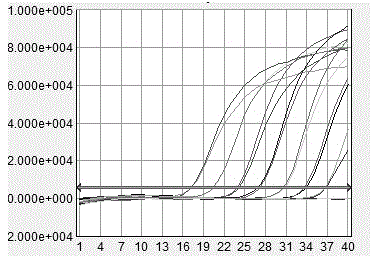
[0047] Primers and probes for fluorescent quantitative PCR amplification of three kinds of Chlamydia (including Chlamydia abortus, Chlamydia abortus, Chlamydia psittaci) were designed based on the reference sequences of MOMP genes of three Chlamydia published by GenBank, compared with MEGA5, and analyzed sequence and design primers and probes in its conserved regions. The probe design software Primer Express 3.0 was used to design 12 sets of fluorescent quantitative primers, which were synthesized by Yingjun (Shanghai) Co., Ltd. ABI 7500 FAST PCR instrument was used to monitor the amplification in the reaction process in real time. The starting time of the amplification, the time to enter the maximum amplification rate, the maximum amplification rate and the time required to reach the plateau were analyzed, and three groups of fluorescent quantitative PCR amplification with the highest amplification rate and good specifici...
Embodiment 2
[0059] Embodiment 2, the preparation of positive control substance
[0060] The kit was used to extract the nucleic acid of the cultured cells of Chlamydia abortus, the nucleic acid was identified by PCR and electrophoresis, and PCR upstream primer SEQ ID NO.10 and PCR downstream primer SEQ ID NO.11 were used to amplify according to the conventional PCR amplification method, The amplified bands were recovered using a gel extraction kit. According to the ratio of 1:10 and the pMD19T vector, the ligation reaction was carried out, ligated overnight at 4°C, and transformed into DH5α bacteria. After resistance selection and PCR identification were positive, it was sequenced and verified, and the OD value of its nucleic acid was measured by a spectrophotometer to make it 260 The ratio of / 280 is between 1.8 and 2.0. Obtain positive recombinant plasmid DNA of Chlamydia abortus. Domestic animal Chlamydia and Chlamydia psittaci positive recombinant plasmid DNAs were prepared accordin...
Embodiment 3
[0061] Embodiment 3, the preparation of negative control substance
[0062] The DNA of the epithelial tissue without the above three kinds of Chlamydia was extracted with a kit, and identified by PCR and electrophoresis.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com