Novel high-throughput detection method for microbial production for dibasic acid based on coupled enzymatic reaction
A dibasic acid and microbial technology, applied in biochemical equipment and methods, microbial determination/testing, etc., can solve the problems of retention time offset, low sensitivity, expensive NADH, etc., and achieve increased screening speed, high sensitivity, and low cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Dual-enzyme-coupled fluorometric method compared to chemichromogenic method.
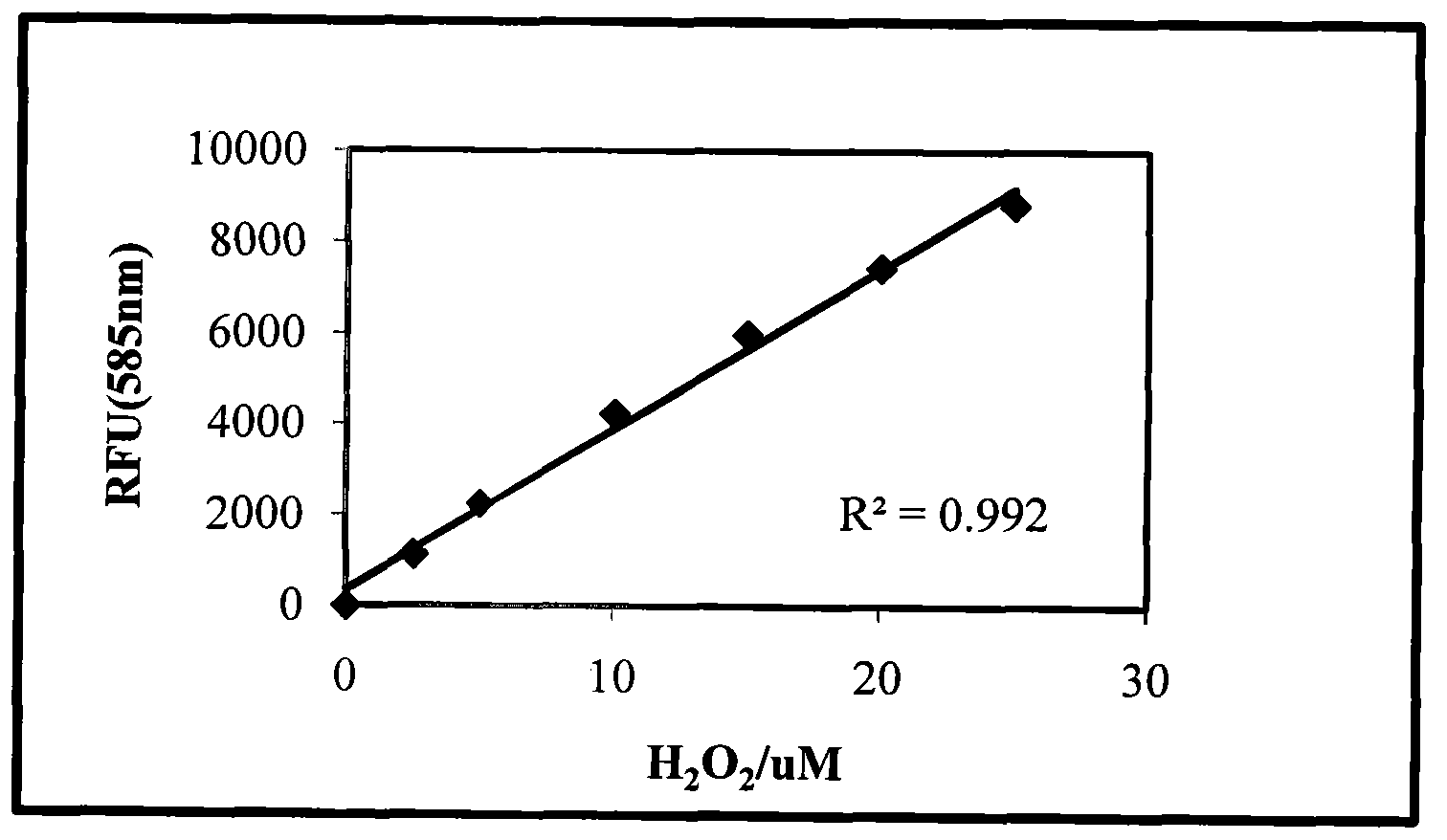
[0029] (1) Fluorescence method. Reagent A: AmplexUltraRed: 0.1 mM, Peroxidase: 9 U / mL, PBS: pH 7.4. Hydrogen peroxide solution: 0.1 mM. Steps: Add different concentrations of H in sequence on a 96-well black microplate 2 o 2 (PBS dilution) 50uL, reagent A 50μL, the total volume is 100μL, measure Ex / Em490 / 585 (nm) fluorescence value. Establish Ex / Em 490 / 585(nm) fluorescence value and H 2 o 2 Concentration standard curve (attached figure 1 ), R 2 =0.992, indicating that the dual-enzyme-coupled fluorescence method can detect μM level of H 2 o 2 .
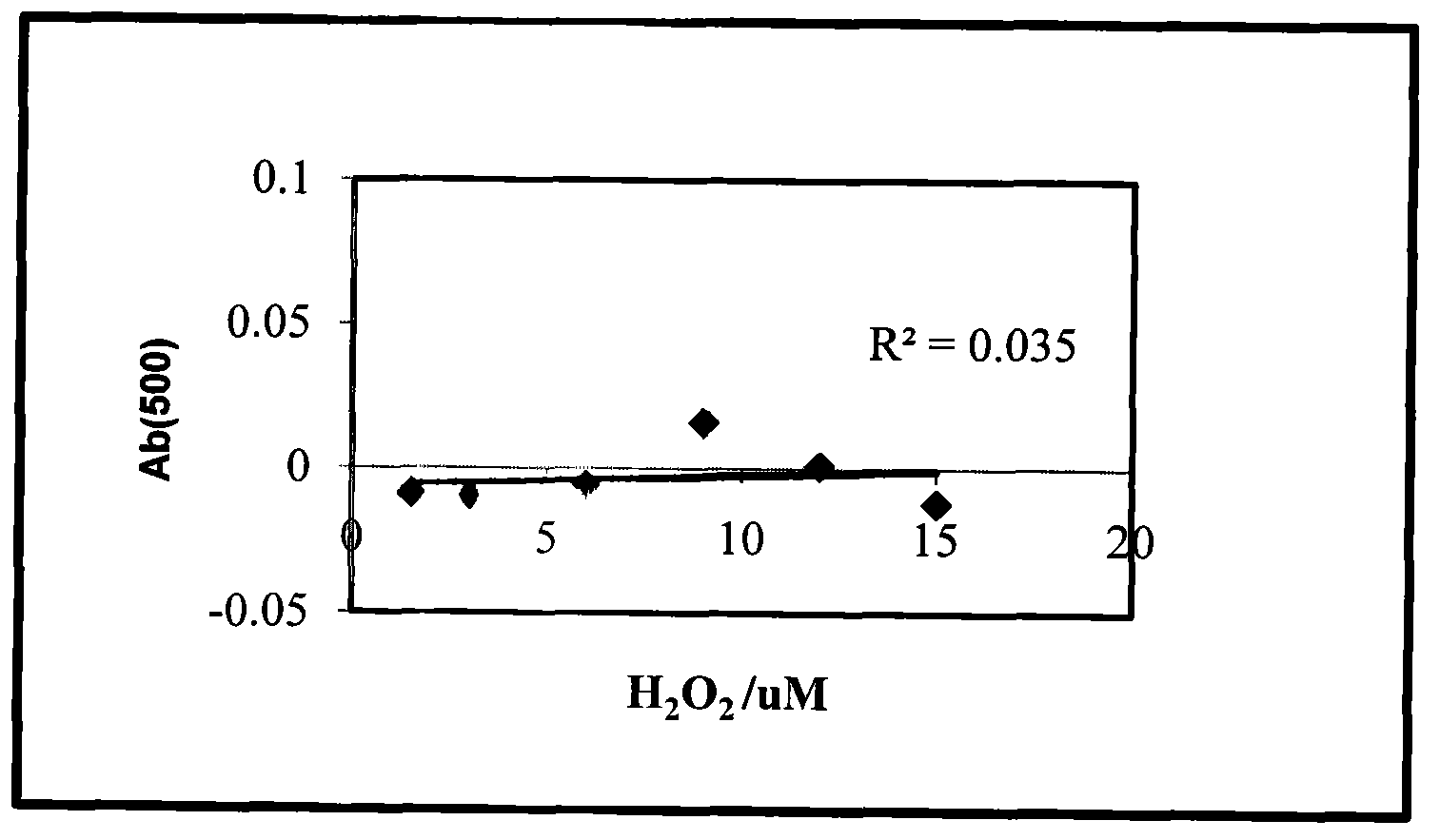
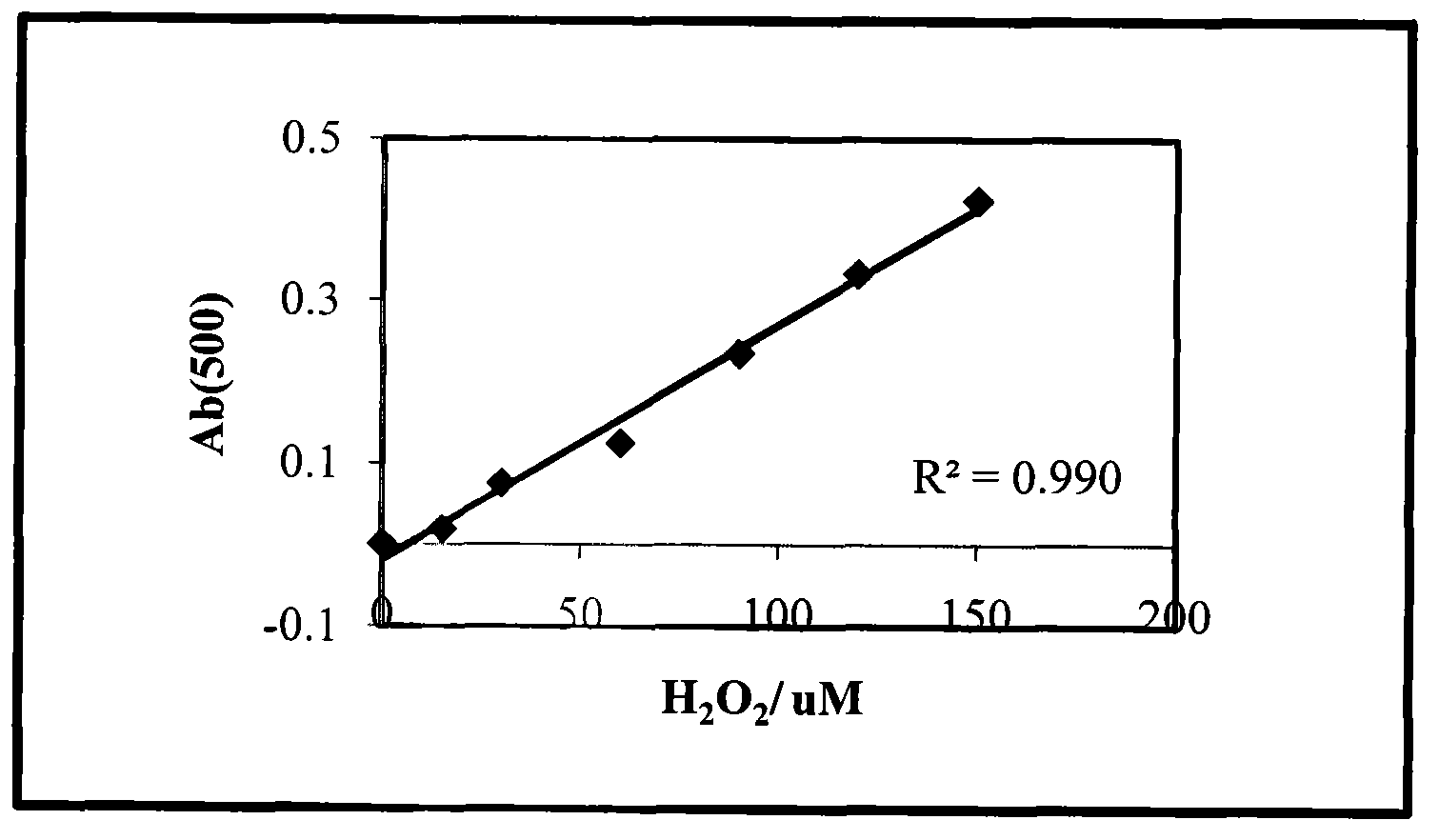
[0030] (2) Chemical chromogenic method. The content of each substance in Buffer I is: phenol: 6mmol / mL, EDTA: 75mg / L, phosphate buffer: pH=7.5, 0.1mol / L. The content of each substance in BufferII is: catalase: 600U / L, 4-antipyrine: 3.5mmol / L, 0.1mol / L phosphate buffer solution pH=7.5. Steps: Add different concentrations of H to the 96-wel...
Embodiment 2
[0032]The method of coupling fumarate reductase and peroxidase is used to measure the change of the fluorescence value of succinic acid with time and the determination of the measurement time. Reagent A: AmplexUltraRed: 0.1 mM, Peroxidase: 9 U / mL, PBS: pH 7.4. Fumarate reductase crude enzyme solution: recombinant Escherichia coli E. coli BL21 (pET-30a-frd) was induced to express and treated with protein extraction reagent to obtain crude enzyme solution. Prepare 1 mM succinic acid solution. Steps: Add 20 μL of PBS, 10 μL of 1 mM succinic acid (diluted in PBS), 50 μL of reagent A, and 20 μL of crude fumarate reductase enzyme solution to a 96-well black microwell plate, with a total volume of 100 μL. The control group did not add succinic acid. Set the temperature of the microplate reader to 37°C, and read the Ex / Em 490 / 585 (nm) fluorescence value every 5 minutes for 0-30 minutes. From the time change graph (attached Figure 4 ), it can be seen that the crude enzyme solution...
Embodiment 3
[0034] The standard curve of succinic acid was determined by the coupling method of fumarate reductase and peroxidase. Reagent A: AmplexUltraRed: 0.1 mM, Peroxidase: 9 U / mL, PBS: pH 7.4. Fumarate reductase crude enzyme solution: recombinant Escherichia coli E. coli BL21 (pET-30a-frd) was induced to express and treated with protein extraction reagent to obtain crude enzyme solution. Prepare 1 mM succinic acid solution. Steps: 30 μL of different concentrations of succinic acid (diluted in PBS), 50 μL of reagent A, and 20 μL of crude fumarate reductase enzyme solution were sequentially added to a 96-well black microwell plate, with a total volume of 100 μL. Incubate at 37°C for 30 minutes, and read the fluorescence value of Ex / Em 490 / 585 (nm). Establish the standard curve of Ex / Em 490 / 585 (nm) fluorescence value and succinic acid concentration (attachment Figure 5 ), R 2 =0.998, indicating that the concentration of succinic acid has a significant linear relationship with the...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com