Method, primer, probe and kit for screening and identifying BCR-ABL unusual fusion types
A very common and fusion technology, applied in the fields of biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., it can solve the problems of false negative, prolonged time period, inability to make clear analysis and judgment, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0120] Extraction of blood RNA: add 1ml of 1× red blood cell lysate to a 1.5ml centrifuge tube, take 0.5ml of the blood sample to be tested, and mix upside down. Centrifuge at 4000rpm for 3min, aspirate the supernatant, add red blood cell lysate and wash once to obtain the required cells; add 1 ml TotalRNA Isolation Reagent (Shanghai Pufei Biotechnology Co., Ltd.), repeatedly pipetting until there are no obvious cell clumps, add chloroform 200μl, vortex and mix for 30s, and let stand on ice for 10min. Then, centrifuge at 4°C at 14,000 rpm for 10 min. Use a pipette to transfer 450 μl of the supernatant to another centrifuge tube, add an equal volume of pre-chilled isopropanol, invert and mix, and then let it stand on ice for 10 min. Centrifuge at 14,000 rpm for 10 min at 4°C. Then use 75% ethanol and absolute ethanol to wash and centrifuge once. Dry at room temperature for 5 minutes, and add 50μl DEPC-H2O to dissolve to obtain RNA extract.
[0121] The formula of 10× red blood...
Embodiment 2
[0123] Reverse transcription: Take 4μl of RNA extract in Example 1 (concentration about 200ng / μl), add 1μl Primer mix (ReverTra AceqPCR RT Kit, purchased from Toyobo (Shanghai) Biotechnology Co., Ltd.) and 3μl DEPC-H2O and mix well. Pre-denaturation at 70℃ for 5min; after quenching on ice for 1min, add 4μl 5*RT buffer (ReverTra Ace qPCR RT Kit), 1μl Enzyme Mix (ReverTra Ace qPCR RT Kit), and add 7μl DEPC-H 2 0 to 20μl total volume. After reacting at 37°C for 60 minutes and then inactivating at 98°C for 5 minutes, the result is the cDNA of the blood sample to be tested.
Embodiment 3
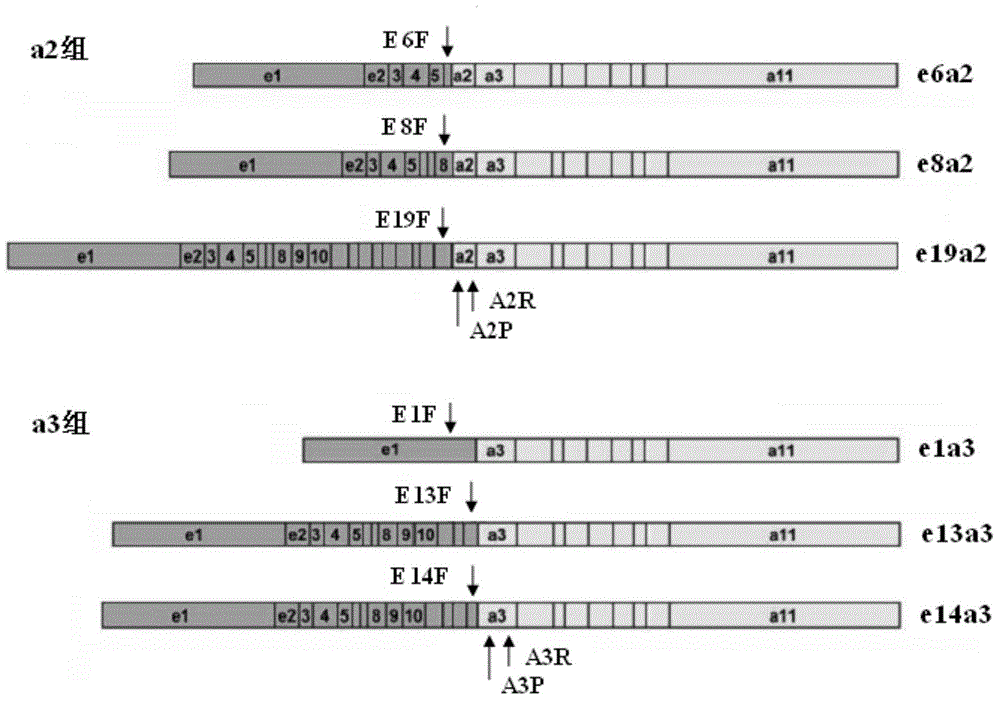
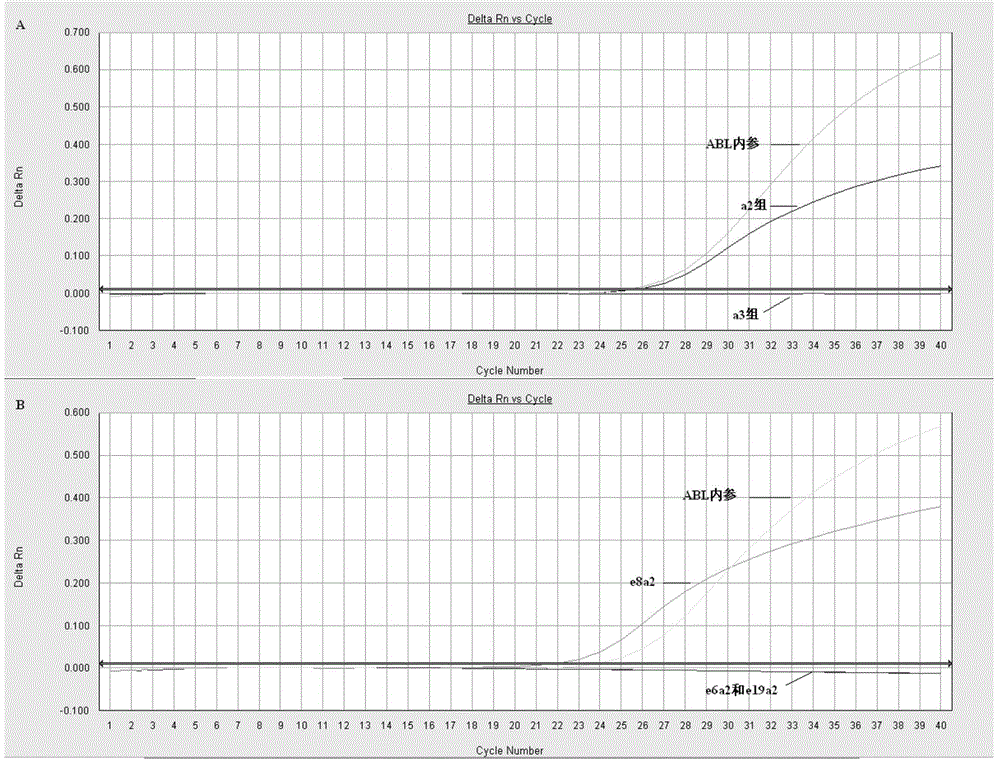
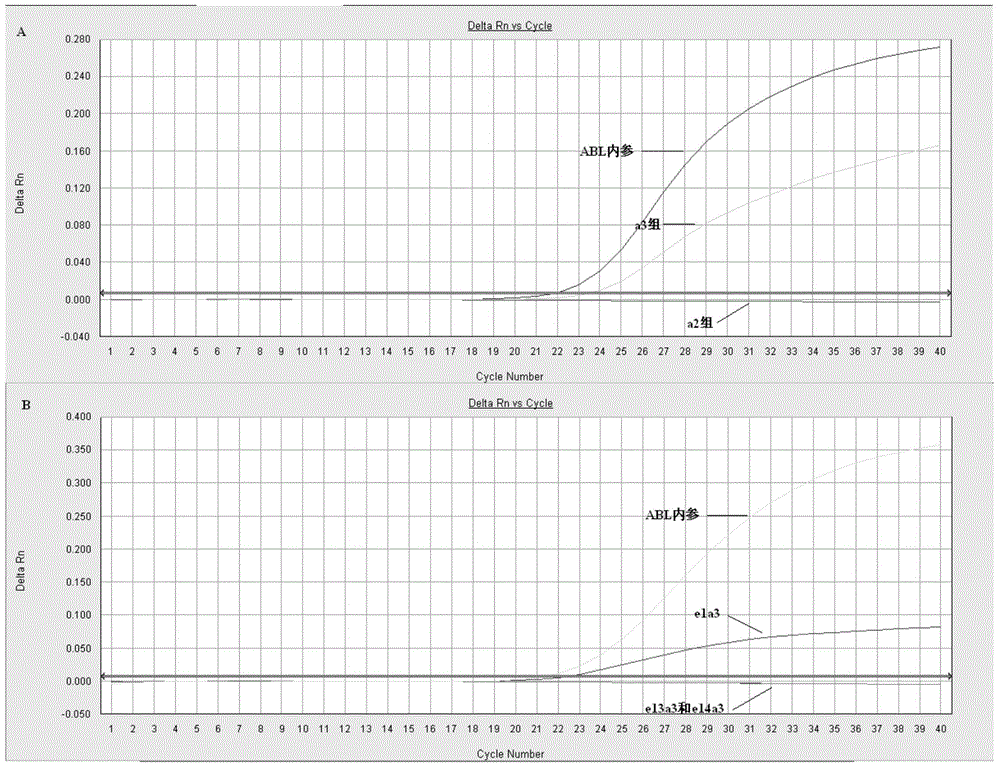
[0125] Fluorescence PCR preliminary screening: configure the preliminary screening reagents according to the materials and dosage shown in Table 1. The preliminary screening reagent contains a total of 3 reaction tubes: a2 group, a3 group and ABL each; ABL is the internal reference detection tube, used to judge whether the RNA extraction quality meets the requirements. 2 μl each of the cDNA obtained in Example 2 was added. The detection was carried out according to the following procedure: 95°C pre-denaturation for 30s; 95°C for 10s, 58°C for 35s, 40 cycles in total; fluorescence was collected at 58°C. HUNDERBIRD Probe qPCR Mix was purchased from Toyobo (Shanghai) Biotechnology Co., Ltd.
[0126] Table 1
[0127]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com