Pyrophosphoric acid sequencing method combined with fluorogenic quantitative PCR technology
A technology of pyrosequencing and fluorescence quantification, applied in the field of pyrosequencing, can solve the problems of sequencing primer consumption, low sensitivity of electrophoresis detection, difficult quantification, etc., and achieve the effect of reducing the risk of false positives, avoiding a large amount of loss, and reducing the rate of missed detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1: Design and synthesis of primers and probes
[0038] Aiming at the 12 and 13 codon mutations of the human KRAS gene, specific PCR primers, fluorescent labeling probes and sequencing primers were designed. The sequences are shown in Table 1, and Sangon Bioengineering (Shanghai) Co., Ltd. was commissioned to synthesize and label them.
[0039] Table 1
[0040]
Embodiment 2
[0041] Embodiment 2: Preparation of DNA template
[0042] Paraffin-embedded sections (10um thick, 1x1 cm 2 ) 2 slices, human tumor tissue DNA was extracted from the slices using a paraffin-embedded tissue genome extraction kit (Tiangen Biochemical Technology (Beijing) Co., Ltd., product number: DP331), which was recorded as sample 1 (case sample).
[0043] Take 500ul of whole blood from a healthy person, and use a blood genome extraction kit (Tiangen Biochemical Technology (Beijing) Co., Ltd., product number: DP318) to extract human genomic DNA from it, which is recorded as sample 2 (control sample).
Embodiment 3
[0044] Embodiment 3: fluorescent quantitative PCR
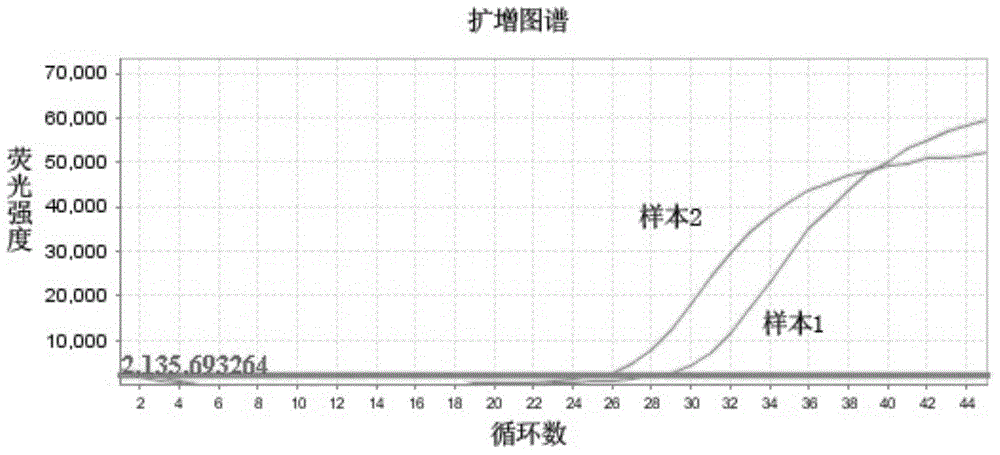
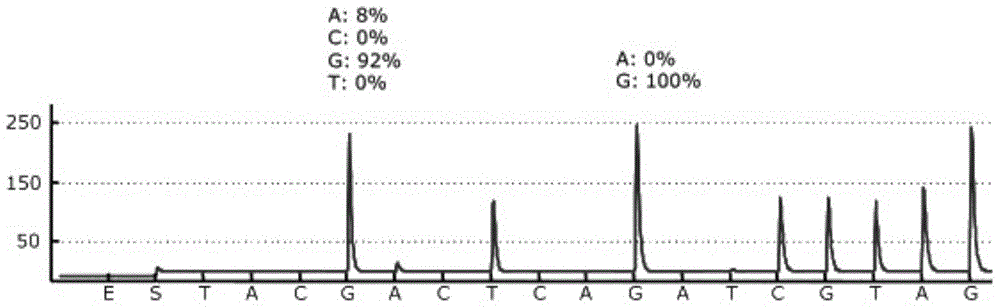
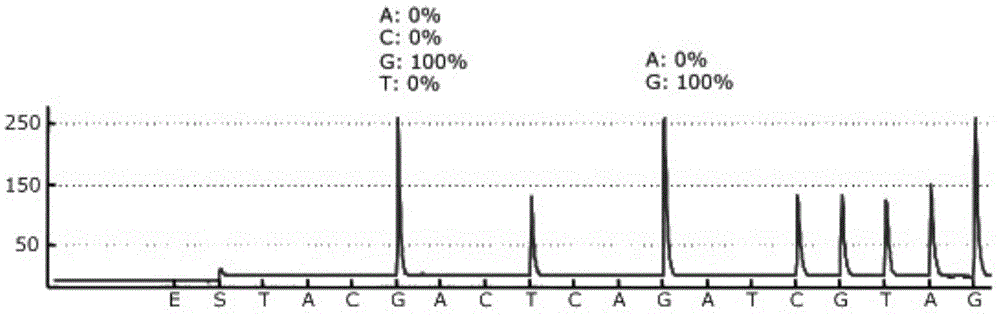
[0045] Using the ABI7500 fluorescent quantitative PCR instrument, perform fluorescent quantitative PCR on the region where the 12 and 13 codons of the human KRAS gene of sample 1 and sample 2 are located, the results are as follows figure 1 shown.
[0046] The reaction system of fluorescent quantitative PCR is:
[0047] DNA template: the DNA template obtained in Example 2, 2ul;
[0048] PCR primers: each 0.4umol / L (final concentration) of the upstream and downstream primers synthesized in Example 1;
[0049] Fluorescent probe: synthesized in Example 1, 0.12umol / L (final concentration);
[0050] PCR reaction solution: 12.5ul (TaqMan Gene Expression Master Mix, Life Technologies company, catalog number: 4369016);
[0051] Ultrapure water: add up to 25ul.
[0052] The reaction conditions of fluorescent quantitative PCR are:
[0053] 37°C, 2 minutes, 1 cycle;
[0054] 95°C, 5 minutes, 1 cycle;
[0055] 95°C, 15 seconds, 6...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com