Myzus persicae resistance associated CYP6CY3 promoter and activity analysis
A technology related to imidacloprid and resistance of peach aphid, applied in the field of DNA sequence and inducible promoter sequence, can solve the problem of lack of promoter of peach aphid
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: Cloning of Peach Aphid CYP6CY3 Promoter
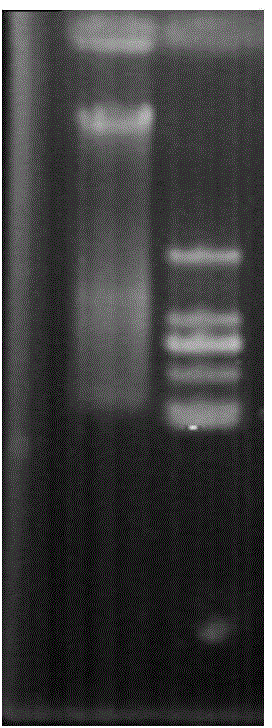
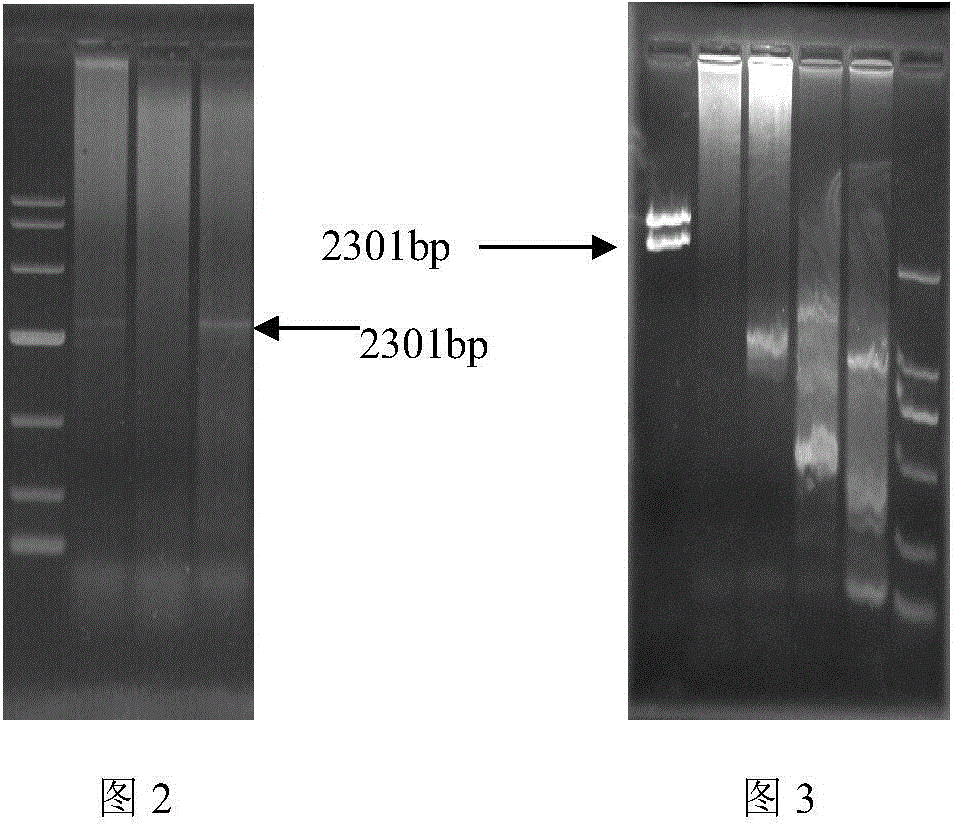
[0025] Chromosome walking primers GSP1 (5-GTCCTCATCTGGAACAGTCCTCCGTAT-3) and GSP2 (5-CAGTCGGTGGTTGACGATGATAGATTC-3) were designed according to the CYP6CY3 mRNA sequence of peach aphid (Genbank No. HM009309). Genomic DNA of green peach aphid ( figure 1 ), and digested with straight cutting enzymes AfeI, EcoRV-HF, PvuII, SmaI, PmeI, StuI, purified DNA and ligated adapter primers. PCR amplification was carried out according to the designed specific primers, and the amplification was carried out according to the Genome walker (Clontech) PCR method. The target fragment was amplified and analyzed by 1% agarose gel electrophoresis, and a band with a length of about 2300bp was amplified ( figure 2 ), and recycle. The recovered fragment was ligated with the pGEM-T vector to obtain a recombinant plasmid, which was extracted and verified by enzyme digestion ( image 3 ), and sequenced.
[0026] Green peach aphid CYP6CY3 pr...
Embodiment 2
[0029] Example 2: Construction of the expression vector pGL3-CYP6CY3(-2230 / +71) of peach aphid CYP6CY3 promoter
[0030] The promoter and 5' untranslated region (see sequence listing) of peach aphid CYP6CY3 cloned in Example 1 were inserted into the pGL3 vector, thereby constructing the pGL3-CYP6CY3(-2230 / +71) vector.
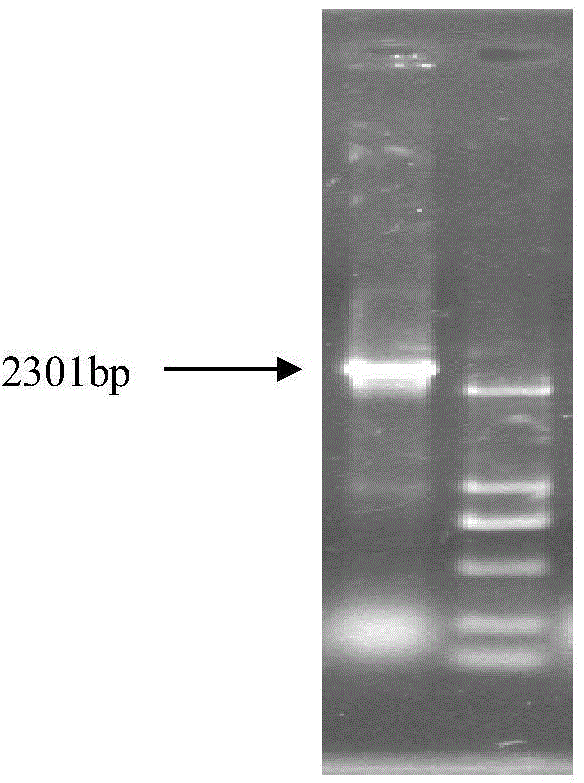
[0031] More specifically, the promoter sequence was amplified using primers (5-ACGCGTATTTATGACTATAACGAGG-3, 5-CTCGAGCAGTCGGTGGTTGACGATGAT-3) and cloned into pGL3 vector MluI and XhoI sites to construct pGL3-CYP6CY3(-2230 / +71 ), used to drive the expression of Luc+, identified by PCR ( Figure 4 ) and enzyme digestion identification ( Figure 5 ) to obtain the recombinant plasmid of the promoter and the vector.
Embodiment 3
[0032] Embodiment 3: Identification of the activity of the promoter of CYP6CY3 of peach aphid according to the present invention
[0033] Sf9 cells were seeded in 24-well plates (4×10 5 cells / well), the pGL3-CYP6CY3 (-2230 / +71) construct (2 μg / well) and the control reporter gene plasmid phRL-TK (Promega; 0.2 μg) were transiently co-transfected with CellfectinII reagent (Invitrogen; 2 μL / well). / hole),. After 48 hours, cells were harvested and the resulting lysates were used to measure luciferase activity (Promega). Such as Figure 6 As shown, pGL3-CYP6CY3 (-2230 / +71) transfected cells have very high luciferase activity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com