Method for identifying whether anammox bacteria in anammox reactor sludge utilize organic matter
A reactor and organic matter technology, applied in the field of identifying whether microorganisms in the sludge of ANAMMOX reactors utilize organic matter, can solve the problems that affect the accuracy of the results, cannot meet the needs of ANAMMOX bacteria research, and the experimental results lack credibility and convincing.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
[0024] Specific implementation mode one: the identification method of this implementation mode is carried out according to the following steps:
[0025] One, use 15 N labeled free ammonia in the ANAMMOX reactor ( 15 NH 4 + -N), use 13 C marks the organic matter in the ANAMMOX reactor, then reacts with anaerobic ammonium oxidation for 10 to 100 hours, then terminates the reaction, centrifuges to obtain microbial samples, and places the microbial samples in 2.5% glutaraldehyde phosphate buffer for fixation;
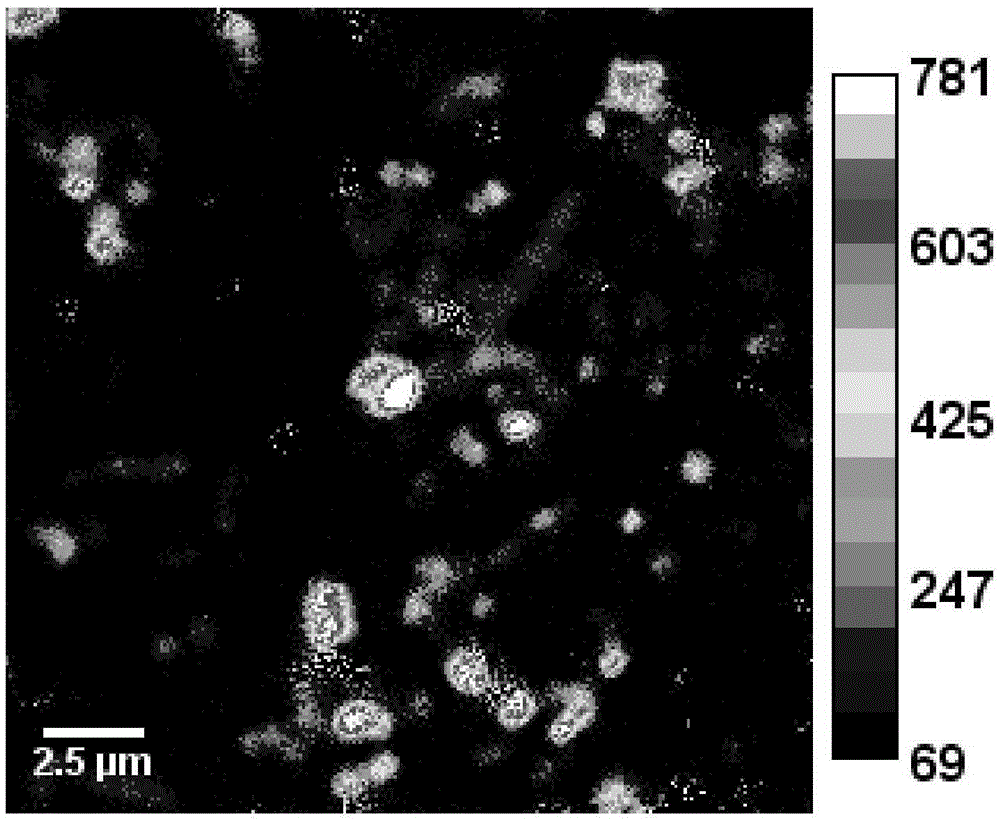
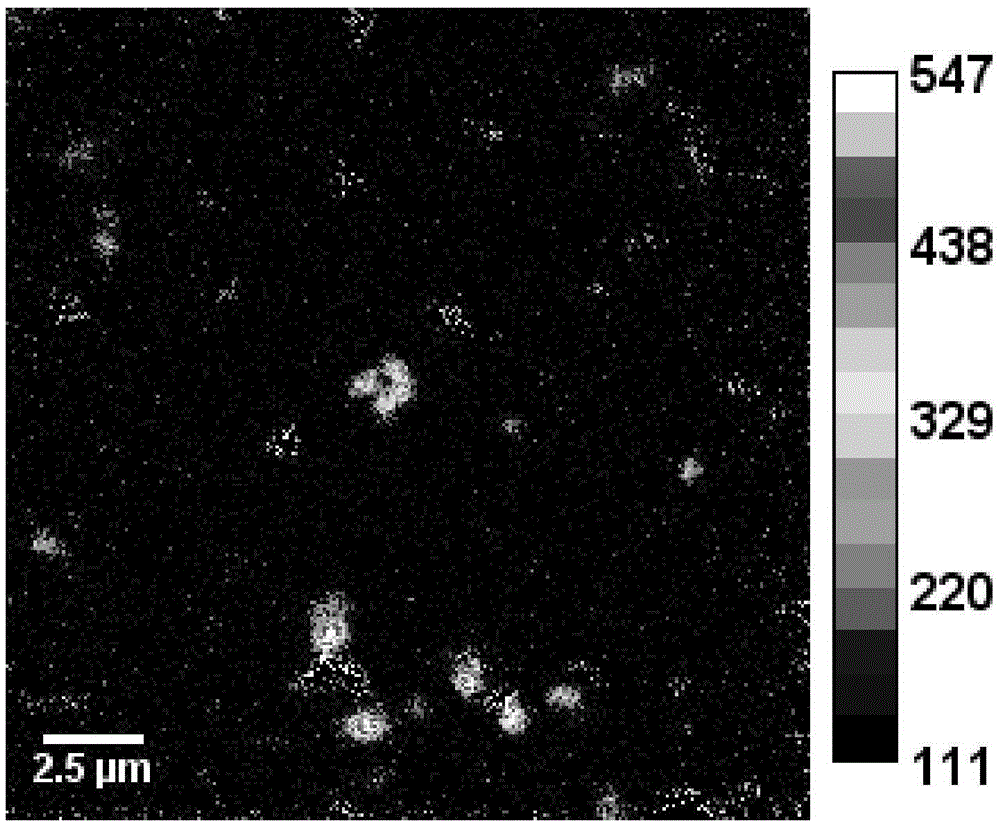
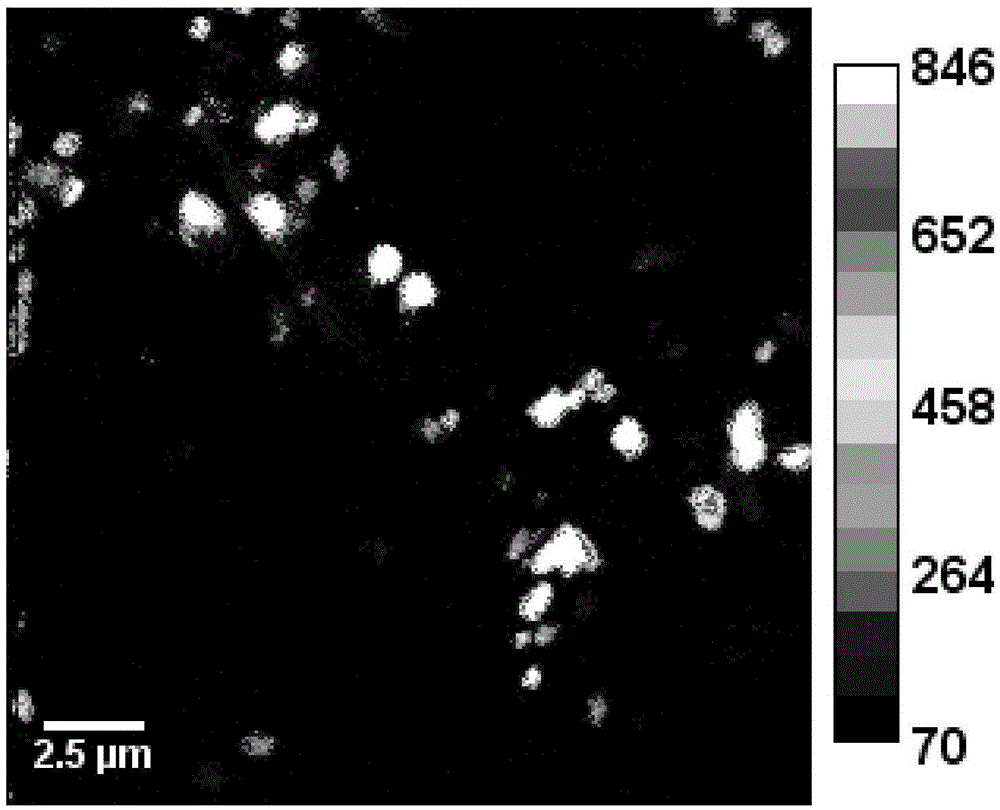
[0026] 2. Prepare a transmission electron microscope sample (TEM sample) with the microbial sample in step 1, and then perform TEM observation;
[0027] 3. According to the TEM observation results of step 2, select the position where the target microorganisms are evenly distributed, use a microtome to slice into semi-thin slices, place them on a conductive silicon wafer, and spray gold again after fixing and drying to obtain a conductive sample;
[0028] 4. Use nano-se...
specific Embodiment approach 2
[0032] Specific embodiment two: the difference between this embodiment and specific embodiment one is: step two transmission electron microscope samples are prepared according to the following steps:
[0033] A. Take out the sample from 2.5% glutaraldehyde phosphate buffer solution, rinse it three times with 0.1mol / L phosphoric acid rinse solution, fix it with 1% osmic acid fixative solution for 2-3 hours, and then rinse it with 0.1mol / L phosphoric acid solution Rinse three times, each time for 15 minutes; then dehydrate the sample with ethanol solutions with gradient concentrations of 50%, 70%, 90%, and 95% at 4°C, and treat each concentration for 15 minutes; then use The ethanol-acetone mixture and 90% acetone solution were dehydrated three times, each time for 15-20 minutes, and then placed in 100% acetone at room temperature for three times, each time for 15-20 minutes;
[0034] B. Use the mixture of pure acetone and embedding solution to treat the sample treated in step A...
specific Embodiment approach 3
[0040] Embodiment 3: The difference between this embodiment and Embodiment 2 is that the slice thickness in step C is 50-60 nm. Other steps and parameters are the same as those in Embodiment 2.
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
| thickness | aaaaa | aaaaa |
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com