Identification method and special primers for major cold tolerance genes in rice
A technology of main effect gene and identification method, which is applied in the field of biomolecular markers, can solve the problems of large errors and easy false positives, and achieve the effects of high cost, shortening the breeding cycle, and saving production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1: Rice Hybridization and Molecular Marker Analysis
[0036] (1) Construction of chromosome substitution lines
[0037] The cold-tolerant donor parent is the Guangxi common wild rice core germplasm material DP30, and the recipient parent is the sequenced indica variety 9311. Through backcrossing and SSR marker-assisted selection, a chromosome segment substitution line of common wild rice was established. F 1 Continuous backcross with 9311 to BC 2 f 1 , in BC 2 f 1 Begin to use 325 polymorphic SSR markers to do genotype analysis on 10 individual plants of each line, to BC 4 f 1 226 lines containing DP30 fragments were obtained, and 96 pairs of markers evenly distributed on the 12 rice chromosomes were selected for background screening, and the individual plants with a recovery rate of more than 90% were selfed and backcrossed with 9311 to obtain shorter Import the substitution system for the segment. Analyze BC 4 f 1 genotypes, self-crossed individual ...
Embodiment 2
[0045] Example 2: Identification of cold tolerance of parents and targeted populations
[0046] The cold tolerance of the donor parent DP30, the cold-tolerant control Fujisaka 5 and the recipient parent 9311 were identified at the seedling stage. The rate of viable seedlings of 9311 as the recipient parent, the perceptual control, was 6.7%. The rates of viable seedlings of the two donor parents DP30 and the cold-tolerant control Fujisaka No. 5 were 85.7% and 88.5%, respectively. The results are shown in Table 1.
[0047] Table 1 The cold tolerance of parents and controls
[0048]
[0049] Using the established 230 BCs covering the whole genome 4 f 2 The cold tolerance level (live seedling rate) of each family was continuously distributed, and the cold tolerance of the DC907 family was the strongest at 83%, while the lowest was 0. The DC907 family with strong resistance was selected to construct a QTLs marker mapping population, and 332 individual plants were identified for...
Embodiment 3
[0057] Example 3 Verification of Molecular Markers
[0058] Negative varieties of materials used: 80 cold-intolerant materials in the breeding combination of cold-intolerant variety 9311 and DP30×9311.
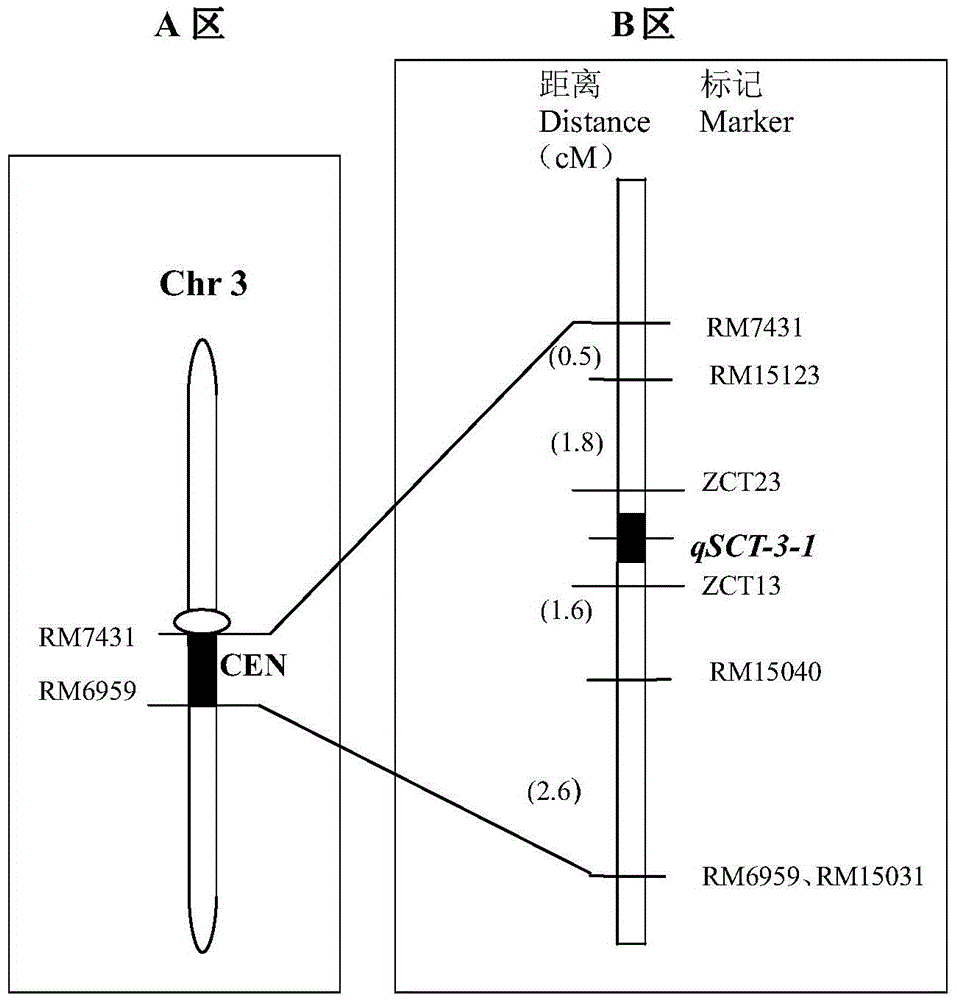
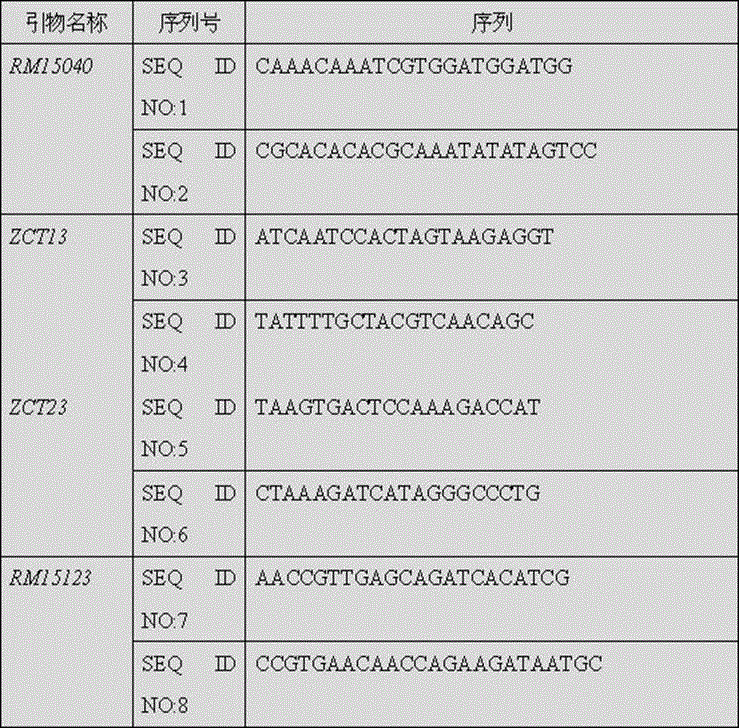
[0059] Positive varieties: 19 cold-tolerant materials in the breeding combination of cold-tolerant parents wild rice DP30 and DP30×9311. Molecular marker primers: RM15040, ZCT13, ZCT23, RM15123.
[0060] Verification method 1: CTAB extraction method to extract genomic DNA from samples of positive rice (rice with the main cold tolerance gene qSCT-3-1) and negative rice (rice without the main cold tolerance gene qSCT-3-1) (the method is the same as the implementation Example 1), using labeled primer RM15040 to amplify sample DNA. Forward primer (SEQ ID NO: 1) 0.10 μM, reverse primer (SEQ ID NO: 2) 0.10 uM, 250 μM dNTPs, 1.0 ul of 10×PCR reaction buffer (50 mM KCl, 10 mM Tris-HCl pH8.3, 1.5 mM MgCl 2 ), rice genomic DNA template 20ng, TaqDNA polymerase 1U, supplement the react...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com