Recombinant pichia pastoris engineering bacteria and metabolic recombinant xylanase as well as preparation of metabolic recombinant xylanase
A technology of xylanase and genetically engineered bacteria, applied in genetic engineering, recombinant DNA technology, enzymes, etc., can solve problems that have not been fully considered
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Embodiment one: the genetically engineered bacteria Pichia pastoris ( Pichia pastoris ) GS115 / pPIC9K-Xyn11Bm The acquisition of the optimized xylanase gene Xyn11Bm of CGMCC No. 9398
[0072] retrieved in GenBank Neocallimastix frontalis Xylanase gene sequence (accession number: AY131336.1), the codons were adjusted and optimized according to the following method:
[0073] (1) Initially replace the codons of Xyn11B according to the yeast codon preference.
[0074] (2) Use Oligo 6.0 to analyze the restriction endonuclease cleavage sites in the gene sequence after replacement, and eliminate the cleavage sites of Bgl II, Sac I, Sal I, EcoR I and Not I in the gene sequence through codon adjustment .
[0075] (3) BioEdit 7.0 was used to analyze the mRNA cryptic splicing sites in the adjusted gene sequence, and the codons were adjusted again to eliminate the cryptic splicing sites GGTAAG, GGTGAT, AATAAA, ATTTA, PolyT and PolyA. And calculate the GC percentage, and then...
Embodiment 2
[0078] Embodiment two: the genetically engineered bacteria Pichia pastoris ( Pichia pastoris ) Construction of GS115 / pPIC9K-Xyn11Bm CGMCC No. 9398
[0079] will be synthesized Xyn11Bm Gene and Pichia pastoris expression vector pPIC9K Eco R I and not I double enzyme digestion, recovery and purification of enzyme digestion products. The purified target gene and expression vector were ligated with T4 DNA ligase, transformed into Escherichia coli DH5α, and the recombinant expression vector pPIC9K- Xyn11Bm . use Sal I vs. pPIC9K- Xyn11Bm Linearize and transform Pichia pastoris GS115 competent cells by electroporation. The transformed bacteria solution was spread on MD plates and incubated at 28-30°C for 48 h. Colonies with normal growth were selected for methanol utilization phenotype identification. The clones with correct methanol utilization phenotype were screened with geneticin G418 sulfate to obtain recombinant yeast engineering bacteria with high copies o...
Embodiment 3
[0080] Example 3: Preliminary screening of xylanase activity of recombinant Xyn11Bm Pichia pastoris
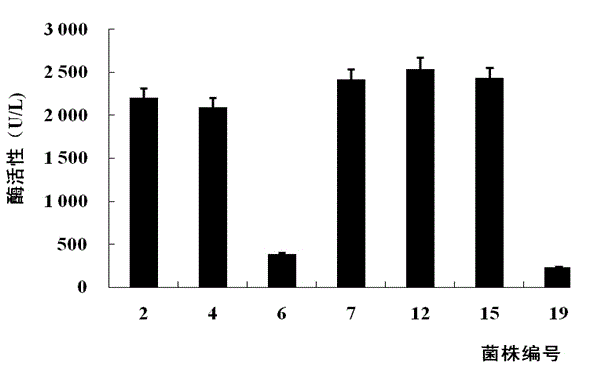
[0081] Inoculate 7 strains resistant to 4 mg / mL Geneticin G418 into 50 mL test tubes containing 5 mL of BMGY medium, culture with shaking at 29°C for 24 hours, then centrifuge and resuspend in BMMY medium, add methanol every 24 hours to make the end point The concentration was kept at 0.5%, and the culture was induced for 48 h. After the cultivation, the supernatant was collected by centrifugation and the enzyme activity was determined with oat xylan as the substrate.
[0082] The enzymatic activities of the 7 strains are shown in the appendix figure 2 shown. Seven strains resistant to 4 mg / mL Geneticin G418 all had xylanase activity, the highest of which was No. 12 (named Xyn11Bm-12), which was 2541U / mL.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Enzyme activity | aaaaa | aaaaa |
| Specific vitality | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com