Recombinant pichia pastoris for heterogenous high level expression of lipase
A lipase and recombinant vector technology, applied in recombinant DNA technology, fermentation, hydrolase and other directions, can solve the problems of high enzyme cost and low enzyme activity, achieve high expression efficiency and reduce separation and purification costs.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] The preparation of embodiment 1 Rhizomucor miehei cDNA
[0026] 1.1 Extraction of Rhizomucor militaris total RNA
[0027] (1) Take an appropriate amount of Rhizomucor miehei (Rhizomucor miehei) mycelium, filter paper to dry up the water, grind with liquid nitrogen, add 1ml Trizol reagent (Invitrogen), oscillate for 5 minutes, and let stand at room temperature for 1 minute;
[0028] (2) Add 0.2ml chloroform, shake for 15s, and let stand for 2min;
[0029] (3) 4°C, 12000rpm, 15min;
[0030] (4) Aspirate the supernatant, add an equal volume of isopropanol, and precipitate at -20°C for 30 minutes;
[0031] (5) 4°C, 12000rpm, 15min;
[0032] (6) Pour off the supernatant, wash the precipitate with 1ml 75% ethanol, 7500rpm, 4°C, 5min;
[0033] (7) Repeat step (6) once;
[0034] (8) Pour off the supernatant and dry for 10 minutes;
[0035] (9) Add appropriate amount of DEPC water to dissolve to obtain total RNA;
[0036] 1.2 Preparation of the first strand of Rhizomucor ...
Embodiment 2
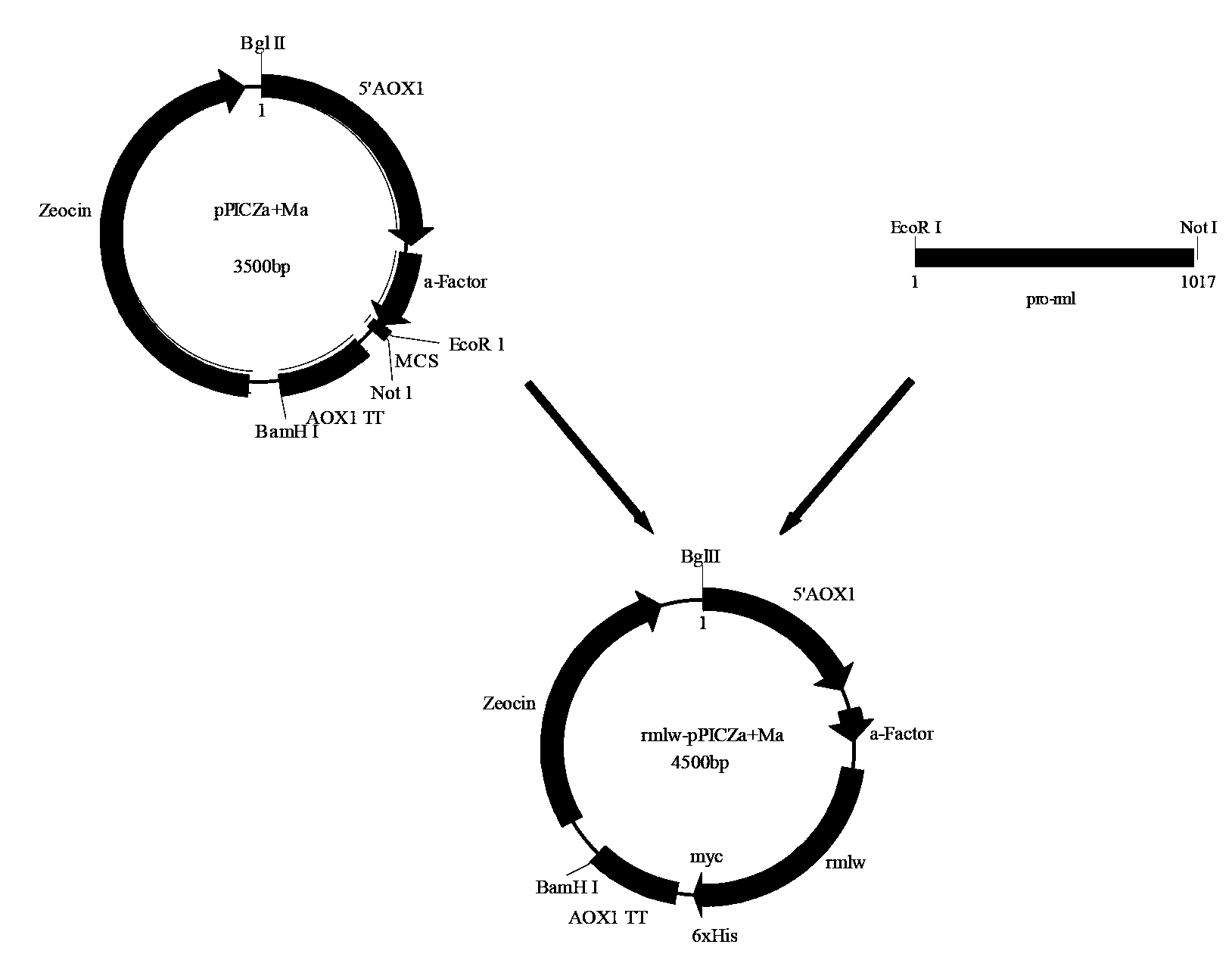
[0041] Example 2 The acquisition of the Rhizomucor miehei lipase gene pro-rml containing the leader peptide and the construction of the expression plasmid
[0042] 2.1 Primer design
[0043] According to the sequence of rml gene in GenBank (GenBank accession number is A02536.1), the following pair of primers were designed and synthesized:
[0044] FW (P1): 5'—CG GAATTC GTGCCAATCAAGAG—3'
[0045] REV (P2): 5'-TAG TCTAGA GTACAGAGGCCTGTG—3'
[0046] EcoRI and NotI restriction sites are designed at both ends of P1 and P2 respectively (see the italicized and underlined part in the above sequence)
[0047] 2.2 PCR amplification of Rhizomucor miehei lipase pro-rml containing leader peptide
[0048] Using P1 and P2 primers, using the above-mentioned Rhizomucor miehei cDNA as a template, the PCR reaction system is:
[0049]
[0050] The reaction conditions are: 95°C for 5 minutes; 5°C for 40s, 60°C for 40s, 72°C for 1 min, 30 cycles; 72°C for 10 minutes; 4°C for 2 minutes. ...
Embodiment 3
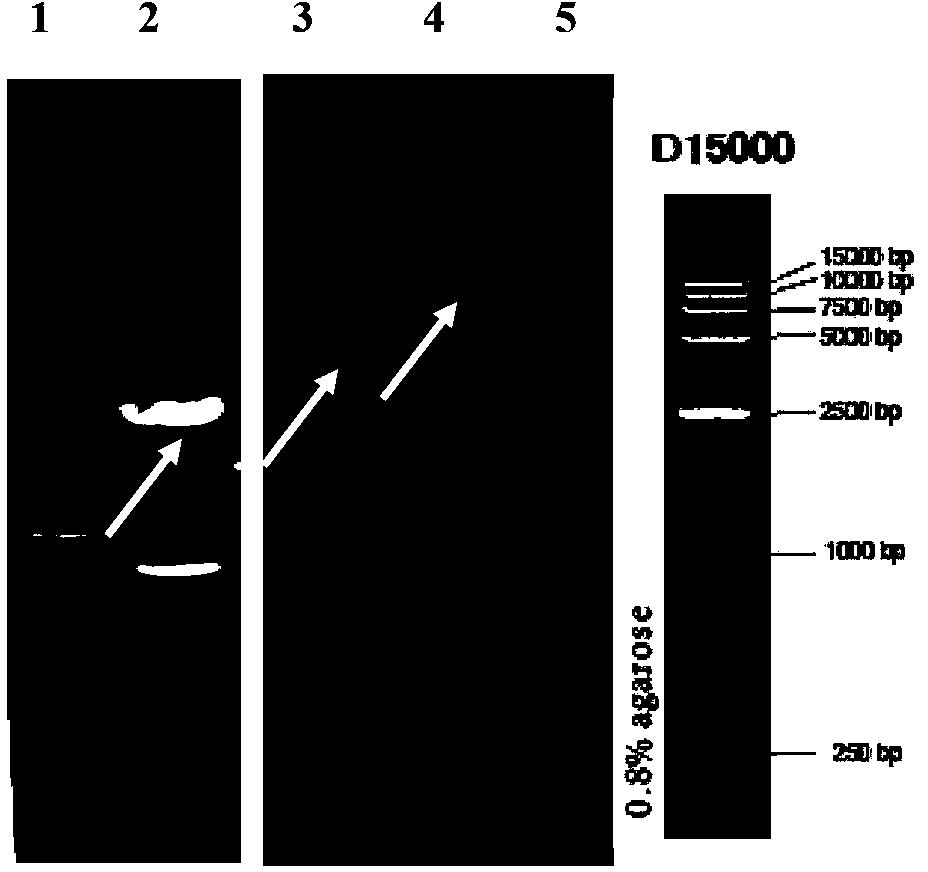
[0061] Example 3 Secretory expression of Rhizomucor miehei lipase gene pro-rml containing leader peptide in Pichia pastoris X-33
[0062] 3.1 Preparation of Pichia pastoris X-33 (purchased by Invitrogen) electroporation competent cells and their electroporation transformation
[0063] (1) Pick a fresh single colony and put it in 5ml YPD liquid medium, and culture it at 30°C, 250rpm for 12-14 hours;
[0064] (2) Inoculate 0.1% of the inoculum into a 2L Erlenmeyer flask containing 500ml of YPD medium, and culture at 30°C, 250rpm for 12-14 hours to make OD600=1.3-1.5;
[0065] (3) Centrifuge at 1500 rpm for 5 minutes at 4°C to collect cells;
[0066] (4) Wash the cells twice with 500-250ml ice-cold sterile water;
[0067] (5) Wash the cells once with 20ml of ice-cold 1M sorbitol solution;
[0068] (6) Resuspend the cells with 1ml of ice-cold 1M sorbitol solution to a final volume of about 1.5ml, and dispense 80μl into small centrifuge tubes;
[0069] 3.2 Electric shock transf...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com