Recombinant adenovirus carrying CAS9 and its application
A recombinant adenovirus and recombinant lentivirus technology, applied to the recombinant adenovirus carrying CAS9 and its application field, can solve the problems of low efficiency, time-consuming and laborious, mutation, etc., and achieve the effects of high efficiency, low cost and fast speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1, the preparation of sgRNA virus liquid and CAS9 virus liquid
[0032] 1. Selection of target sites
[0033] CCAAT-enhancer binding protein-A (CCAAT-enhancer binding protein-alpha, CEBPA) was used as the target sequence to verify the reliability and authenticity of the CRISPR / Cas9 system. In order to avoid off-target effects, two gRNA tandem target regions were selected.
[0034] The DNA sequences of the two gRNAs are as follows:
[0035] gRNA1: 5'-GAAAGGACGAAACACC GAAGGCGGCCGGGTCGATGTAGG gttttagagctagaaat-3';
[0036] gRNA2: 5'-GAAAGGACGAAACACC GCACAGCCGACAGCAGGAGAAGG gttttagagctagaaat-3'.
[0037] gRNA1 is shown in sequence 1 of the sequence listing (its target region is the 323-345th nucleotide from the 5' end of sequence 3 of the sequence listing). gRNA2 is shown in sequence 2 of the sequence listing (its target region is the 375th-397th nucleotide from the 5' end of sequence 3 of the sequence listing). The fragment of the gene encoding CEBPA pro...
Embodiment 2
[0072] Embodiment 2, in vitro activity detection
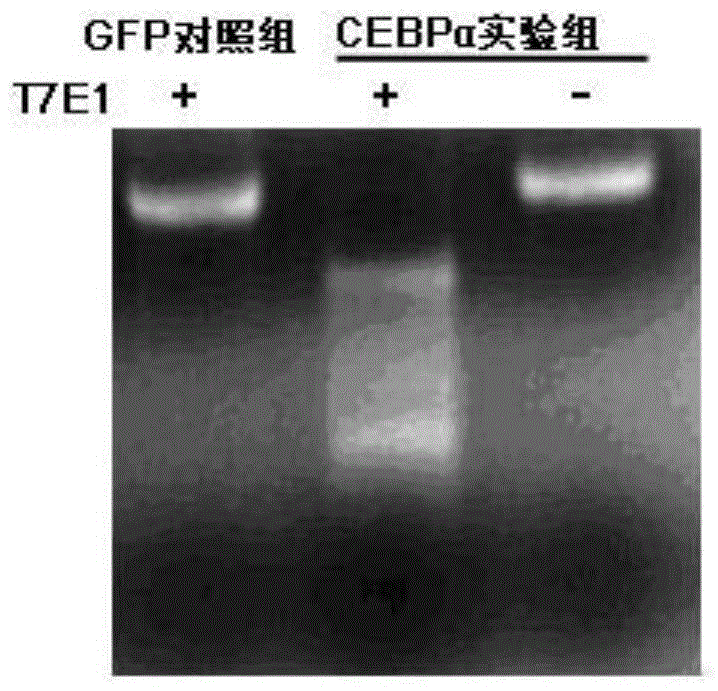
[0073] 1. Suspend AML12 cells in DMEM medium to obtain a cell concentration of 5×10 5 cells / ml of cell suspension.
[0074] 2. Group processing
[0075] Experimental group: Mix 1ml of the cell suspension obtained in Step 1, 1ml of the sgRNA virus solution obtained in Step 3 of Example 1, and 0.1ml of the CAS9 virus solution obtained in Step 5 of Example 1, and mix them at 37°C and 5% CO 2 Cultivate in the incubator for 48h;
[0076] Control group: Mix 1ml of the cell suspension obtained in Step 1, 1ml of the control virus solution obtained in Step 6 of Example 1, and 0.1ml of the CAS9 virus solution obtained in Step 5 of Example 1, and store at 37°C and 5% CO 2 Cultivate in the incubator for 48h.
[0077] 3. After completing step 2, take cells, extract genomic DNA, and perform PCR amplification using the primer pair composed of F1 and R1.
[0078] F1:5'-ATTCGCGACCCGAAGCTGCG-3';
[0079] R1: 5'-TGTTCTTGTCCACCGACTTCTTGGCT-...
Embodiment 3
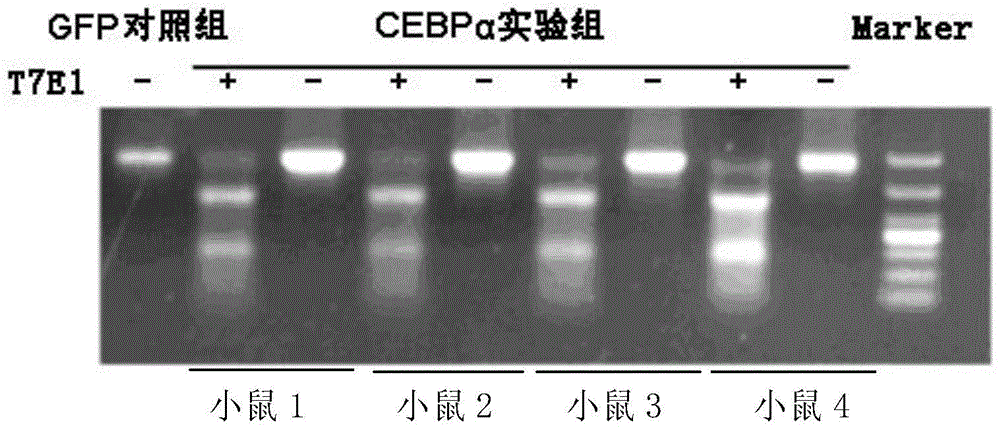
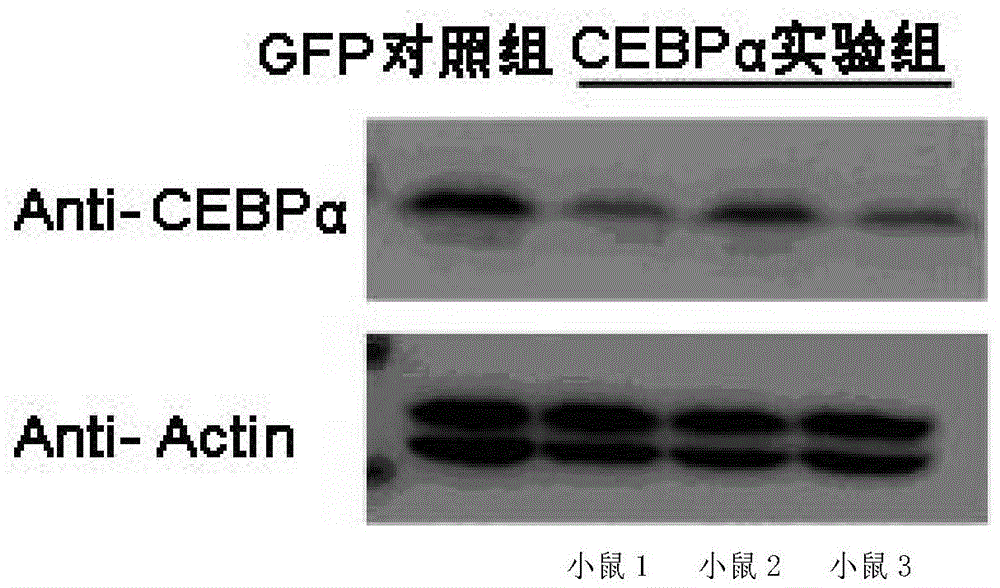
[0081] Embodiment 3, in vivo activity detection
[0082] 1. Group processing
[0083] Experimental group: 4 C57 mice were taken, and each tail vein was injected with 0.1ml of the sgRNA virus solution obtained in Step 3 of Example 1 and 0.1ml of the CAS9 virus solution obtained in Step 5 of Example 1;
[0084] Control group: take 1 C57 mouse, and inject 0.1 ml of the control virus solution obtained in Step 6 of Example 1 and 0.1 ml of the CAS9 virus solution obtained in Step 5 of Example 1 into the tail vein of each mouse.
[0085] 2. Seven days after the injection of the virus solution, the liver of the mouse was taken.
[0086] 3. Take the liver obtained in step 2, extract genomic DNA, use the primer pair composed of F1 and R1 to perform PCR amplification, take the PCR amplification product, and perform T7E1 enzyme digestion (set a control treatment without T7E1 enzyme digestion), and then carry out 2% agarose gel electrophoresis, see the results figure 2 . The control g...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com