Method and application of CRISPR-Cas9 specific knockout of soybean lipoxygenase gene
A lipoxygenase and specific technology, applied in the field of soybean molecular breeding and biology, can solve the problems of time-consuming and laborious, and achieve the effect of avoiding high cost and avoiding long cycle.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1: Construction of CRISPR-Cas9 Gene Knockout Vector and Stable Transformation of Soybean
[0030] Main reagents and sources of materials:
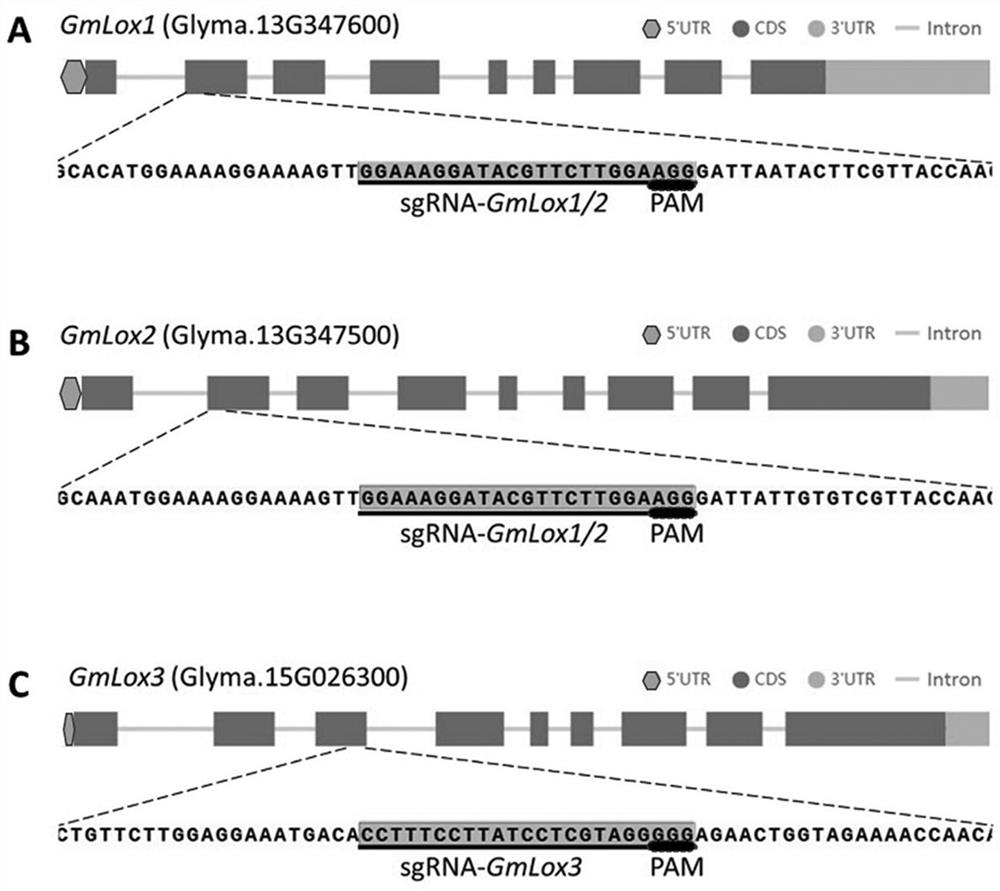
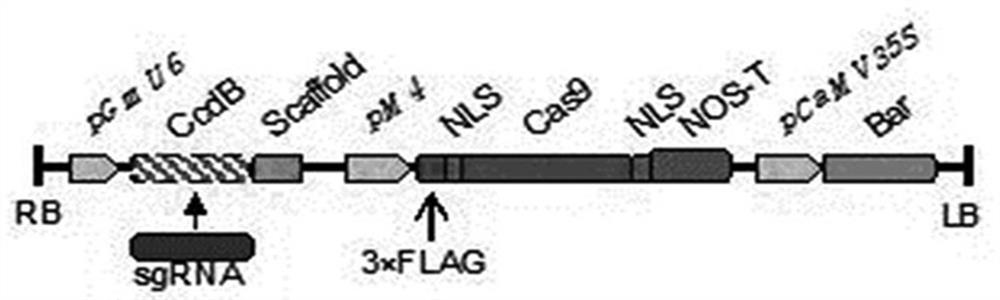
[0031] The Escherichia coli DH5α competent cells, Agrobacterium rhizogenes K599 competent cells and Agrobacterium GV3101 competent cells used in this study were all preserved by our laboratory; the CRISPR-Cas9 gene editing vector pGES201 used was obtained from pCAMBIA1300 ( Cambia, Australia), the pGmU6 promoter promotes the expression of sgRNA, the soybean endogenous promoter pM4 promotes the expression of Cas9, and the empty vector carries its own gene "Escherichia coli toxin-antitoxin system toxin protein ccdB (control of cell division or death system)", and there are enzyme cutting sites BsaI ( figure 1 ).
[0032] PCR Mix enzyme PCR Master Mix was purchased from Qingke Biotechnology Co., Ltd.; BsaI restriction endonuclease was purchased from NEB (UK); T4 ligase was purchased from Beijing Quanshijin Biotechnology Co.,...
Embodiment 2
[0084] Example 2: Separation of T-DNA and Editing Sites GLox Full deletion mutant screening
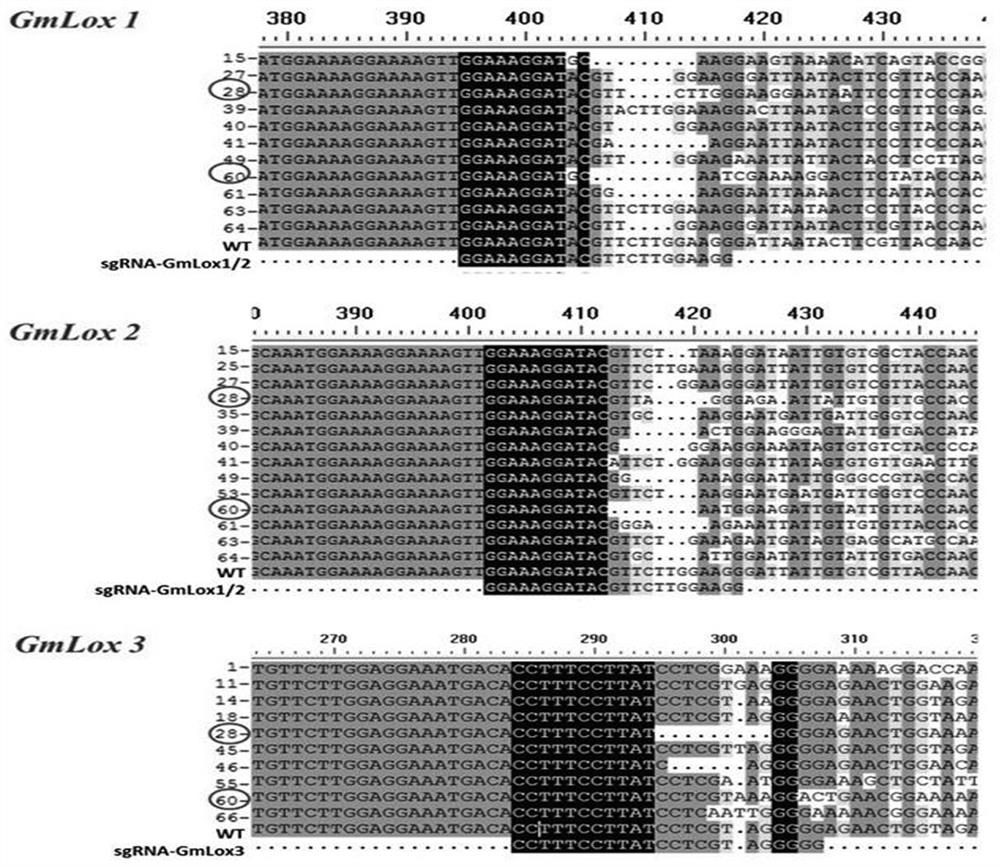
[0085] We used methods such as leaf smearing Basta, Bar Primer PCR (BPP), sgRNA Specific PCR (SSP), and PCR product sequencing to screen positive plants, identify sgRNA distribution, and detect targeted gene editing in stably transformed plants. The results showed that: 76 strains of T 0 Among the stable transformed seedlings, 60 were positive plants, and 27 of them contained only sgRNA- GmLox1 / 2 , 22 strains contained only sgRNA- GmLox3 , 11 strains also contain sgRNA- GmLox1 / 2 and sgRNA- GmLox3 ; 22 positive plants occurred GLox Gene editing (gene editing efficiency is 36.7%), in which positive plants Gmlox -28 and Gmlox -60 knocked out 3 at the same time GLox Gene( image 3 ).
[0086] take T 0 Positive plants Gmlox -28 and Gmlox -60 harvested seeds, planted in pots in the grow room, gain T 1 Substitute plants. take T 1 Extract DNA from leaves of generati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com