Carbonyl reductase gene, codase, vector, engineering bacterium and application thereof
A technology of genetically engineered bacteria and reductase, applied in the fields of genetic engineering, oxidoreductase, application, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
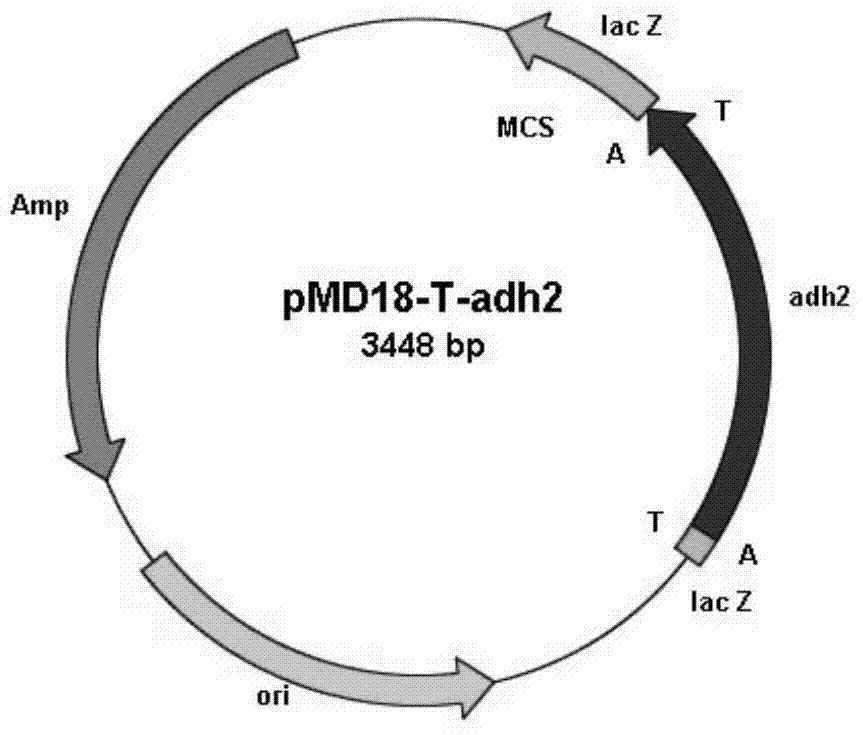
[0045] Example 1: Amplification of the carbonyl reductase gene adh2
[0046] According to the whole genome sequencing information of Burkholderia gladioli (Burkholderia gladioli) ZJB-12126, a large number of carbonyl reductases were excavated, one of which has the ability to catalyze 2-benzamidomethyl-3-ketobutyrate and N,N-Dimethyl-3-oxo-3-(2-thienyl)propionamide generates (2S,3R)-2-benzamidomethyl-3-hydroxybutyrate and (S)-N , N-bismethyl-3-hydroxyl-3-(2-thienyl) propanamide functional enzyme is the carbonyl reductase BgADH2 involved in the present invention.
[0047] Using MPBio's Spin kit extracts the total genomic DNA of Burkholderia gladioli (Burkholderia gladioli) ZJB12126 cells, and uses the genomic DNA as a template to carry out PCR under the action of primer 1 (ATGAGCAAGCGGCTGGAAGGCAAGG) and primer 2 (TCAGACCTGGGCCTGGC CGCC) Amplify. PCR reaction system (total volume 50 μL): 5 μL of 10×Pfu DNA Polymerase Buffer, 1 μL of 10 mM dNTP mixture (2.5 mM each of dATP, dC...
Embodiment 2
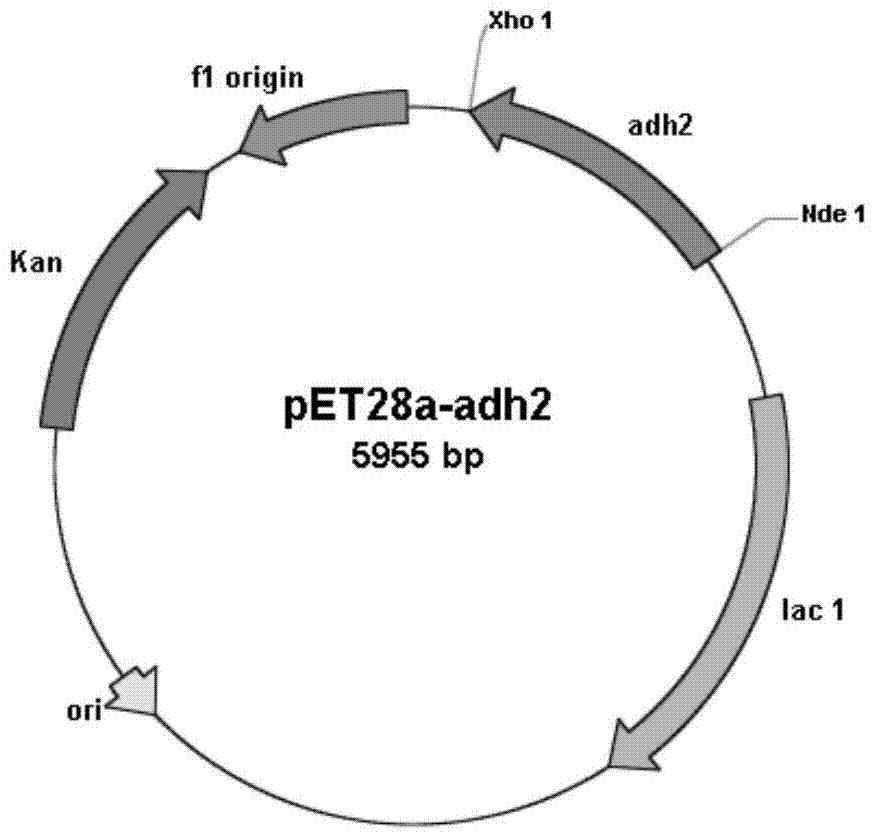
[0050] Embodiment 2: Construction of recombinant Escherichia coli BL21(DE3) / pET28a-adh2
[0051] Design primer 3 according to embodiment 1 analysis result ( CATATG AGCAAGCGGCTGGAAGGCAA GG), primer 4 ( CTCGAG TCAGACCTGGGCCTGGCCGCCG), and Nde I and Xho I restriction enzyme sites (underlined) were introduced into primer 3 and primer 4, respectively. Under the priming of primer 3 and primer 4, high-fidelity Pfu DNA polymerase was used to amplify to obtain a 756bp carbonyl reductase gene sequence (its nucleotide sequence is shown in SEQ ID NO: 1), and after sequencing, use The amplified fragment was treated with Nde I and Xho I restriction endonucleases (TaKaRa), and the fragment was treated with the commercial vector pET28a (Invitrogen) treated with the same restriction enzymes using T4 DNA ligase (TaKaRa). Connection to construct the expression vector pET28a-adh2. The constructed expression vector pET28a-adh2 was transformed into Escherichia coli BL21(DE3) (Invitrogen), sprea...
Embodiment 3
[0052] Embodiment 3: Recombinant carbonyl reductase (BgADH2) wet thallus
[0053] The recombinant Escherichia coli E.coli BL21(DE3) / pET28a-adh2 bacterial cell containing the expression recombinant plasmid pET28a-adh2 obtained in Example 2 was inoculated into LB liquid medium containing a final concentration of 50 μg / mL kanamycin resistance, Cultivate at 37°C for 12 hours at 200rpm, then inoculate with 1% inoculum size (v / v) into fresh LB liquid medium containing kanamycin resistance at a final concentration of 50 μg / ml, and cultivate at 37°C at 150rpm until Cell OD 600 After reaching 0.6-0.8, add IPTG with a final concentration of 0.1mM, induce culture at 28°C for 12h, centrifuge at 5000rpm at 4°C for 5min, discard the supernatant, collect the precipitate, and obtain recombinant Escherichia coli BL21( DE3) / pET28a-adh2 wet cells. The bacterium can be used directly as a biocatalyst or for protein purification.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com